Author: Denis Avetisyan
A new multi-agent system, Indibator, leverages unique research backgrounds to foster more diverse and factually grounded reasoning in the search for novel molecules.
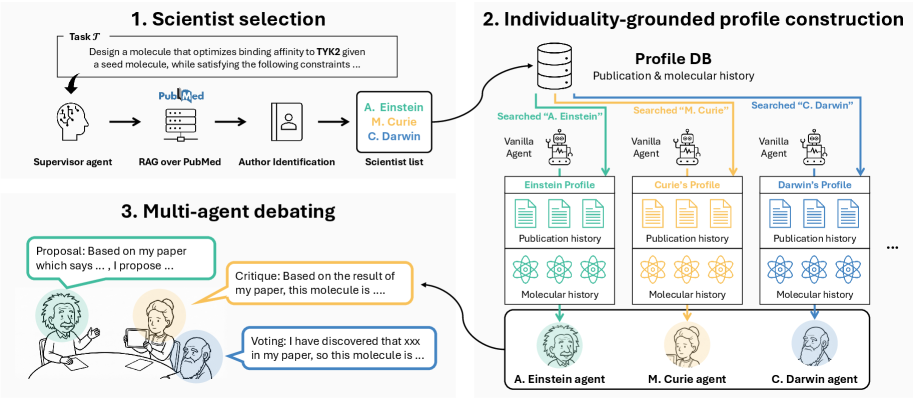
Indibator grounds each agent in a personalized history of publications and molecular data to enhance debate and accelerate scientific discovery.
Current multi-agent systems for scientific discovery often rely on simplified, generic agent personas, failing to capture the nuance of human scientific reasoning. To address this, we present INDIBATOR: Diverse and Fact-Grounded Individuality for Multi-Agent Debate in Molecular Discovery, a framework that grounds each agent in individualized research trajectories derived from both publication history and prior molecular knowledge. Our results demonstrate that these “individuality-grounded” agents consistently outperform those using coarse-grained personas in molecular discovery tasks. Does imbuing agents with a form of “scientific DNA” represent a crucial step toward truly autonomous and creative scientific exploration?
Beyond Chance: Towards Rational Molecular Design
Historically, the creation of new molecules, particularly within pharmaceutical research, has been profoundly shaped by the expertise of chemists and a significant element of chance. This reliance on human intuition, while responsible for many breakthroughs, presents inherent limitations in an era demanding accelerated discovery. The process is intrinsically slow, requiring years of dedicated effort to synthesize and test potential compounds, and increasingly unsustainable given the escalating costs and complexity of modern drug development. Furthermore, serendipitous discoveries, while valuable, are unpredictable and cannot be consistently replicated to meet the growing need for novel therapeutics targeting an expanding range of diseases. This traditional approach struggles to efficiently navigate the vastness of chemical space, hindering the identification of promising candidates that might lie outside the bounds of established chemical knowledge and familiar structural motifs.
Despite advancements in high-throughput screening and computational chemistry, truly innovative drug discovery often stalls due to a limited comprehension of chemical space. These methods, while capable of rapidly evaluating numerous compounds, frequently operate within predefined boundaries, overlooking potentially transformative molecules residing in unexplored regions of this vast landscape. Current algorithms often struggle to accurately predict the properties of compounds significantly different from those already studied, hindering the identification of novel scaffolds and functionalities. Consequently, researchers may unintentionally prioritize incremental improvements to existing drugs over genuinely breakthrough therapies, because the ability to navigate the full complexity of chemical possibilities – encompassing both known and unknown interactions – remains a significant challenge.
Simulating the Scientific Collective: Indibator
Indibator utilizes a multi-agent system architecture to simulate scientific collaboration. Each agent within the system is assigned a distinct ‘Scientific DNA’, representing a unique profile of expertise derived from a combination of factors. This approach moves beyond single-agent optimization by creating a distributed network of specialized ‘researchers’ capable of independently generating and evaluating molecular proposals. The system’s design allows for the emulation of peer review and knowledge sharing inherent in scientific discovery, with agents leveraging their individual profiles to contribute to a collective exploration of chemical space. This distributed architecture is intended to overcome limitations of traditional, centralized approaches to molecular design and discovery.
Each agent within Indibator possesses an ‘Expertise Profile’ derived from two primary data sources: ‘Publication History’ and ‘Molecular History’. The ‘Publication History’ component catalogues an agent’s authored publications, extracting keywords and research foci to establish areas of demonstrated knowledge. Complementing this, ‘Molecular History’ tracks the specific molecules an agent has previously investigated – whether through synthesis, simulation, or analysis – thereby defining their practical experience with chemical structures and properties. This combined profile allows each agent to assess the feasibility and potential impact of proposed molecules, and to formulate critiques based on established scientific precedent and prior investigation, forming the basis for informed contributions to the multi-agent system.
Indibator achieves enhanced chemical space exploration by leveraging the combined expertise of its constituent agents. Traditional methods often rely on singular algorithms or limited datasets, resulting in incomplete searches. In contrast, Indibator’s multi-agent system allows for parallel hypothesis generation and critique, effectively broadening the scope of investigation. Benchmarking studies have demonstrated consistent performance gains over baseline approaches, including random search and single-algorithm optimization, across metrics such as target molecule identification and property prediction. These results indicate that the aggregation of diverse ‘Scientific DNAs’ within Indibator leads to more efficient and comprehensive sampling of the chemical landscape.
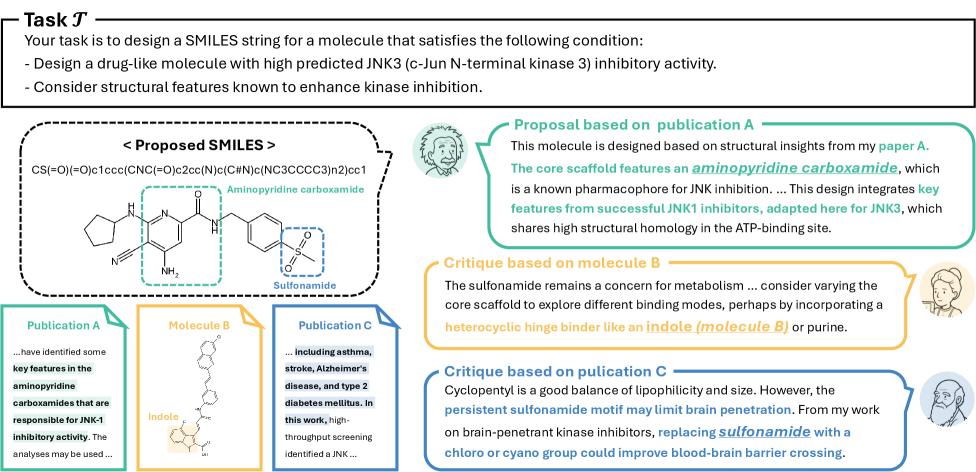
The Logic of Debate: Proposal, Critique, and Selection
The ‘Debating System’ initiates with a ‘Proposal Phase’ wherein individual agents, each possessing specialized expertise in molecular properties and design, independently generate candidate molecules. These agents utilize their distinct knowledge bases – encompassing areas such as chemical synthesis feasibility, predicted bioactivity, and structural characteristics – to propose compounds considered likely to perform well against specified targets. The output of this phase is a diverse set of molecular candidates, representing a broad exploration of the chemical space informed by the collective expertise of the participating agents. This initial diversity is crucial for subsequent evaluation and refinement within the system.
The Critique Phase of the Debating System is characterized by a comprehensive evaluation of proposed molecular candidates. During this phase, individual agents analyze proposals generated in the Proposal Phase, identifying potential weaknesses or limitations in their predicted performance, synthesizability, or novelty. This process involves detailed scrutiny of the rationale behind each proposal and a challenge to underlying assumptions. Agents provide specific feedback, contributing to a rigorous assessment of each candidate’s potential before proceeding to the Voting Phase, ultimately enhancing the quality of the final selection.
The Voting Phase represents the culmination of the Debating System, quantitatively assessing agent critiques to prioritize molecular candidates. This aggregation of feedback utilizes a collective intelligence approach, weighting agent input to determine the most promising molecules for advancement. Performance benchmarks demonstrate substantial improvements – ranging from 17.4% to 123.5% – when comparing molecules selected via this system to those generated using Genetic GFN across a variety of evaluated targets. This indicates the Voting Phase effectively identifies superior candidates through the synthesis of expert evaluation.
![Indibator consistently generates molecules with higher docking scores [latex]\left(representing stronger binding\right)[/latex] and comparable diversity to vanilla debate and keyword persona debate methods.](https://arxiv.org/html/2602.01815v1/x4.png)
Beyond Mimicry: Addressing the ‘Clever Hans’ Effect
A critical challenge in assessing artificial intelligence for molecular discovery lies in avoiding the ‘Clever Hans’ effect – a phenomenon where models appear to perform well not through genuine innovation, but by subtly recognizing the identities of the scientists who designed the molecules in the training data. This can occur if models inadvertently learn to associate specific molecular features with particular researchers, leading to inflated performance metrics on benchmark datasets. To mitigate this bias, researchers have implemented strategies to ensure models focus on the inherent properties of molecules, rather than spurious correlations with their creators. By explicitly addressing this potential pitfall, evaluations become more reliable and accurately reflect a model’s true capacity for generating novel and effective compounds, fostering genuine progress in drug discovery.
Indibator addresses a critical challenge in molecular discovery – the potential for machine learning models to identify researchers based on subtle, unintended biases within benchmark datasets. Rather than allowing agents to freely explore the chemical space, Indibator establishes unique, individualized research trajectories for each agent. This deliberate constraint prevents models from exploiting spurious correlations – patterns that coincidentally link molecular features to the originating scientist – and forces them to genuinely explore novel chemical structures based on target properties. By simulating diverse research paths, the system ensures that apparent ‘creativity’ stems from legitimate innovation, rather than the identification of dataset-specific quirks or researcher fingerprints, ultimately leading to more reliable and robust assessments of a model’s true generative capabilities.
The resulting methodology delivers a more dependable evaluation of molecular design capabilities, evidenced by gains in predictive performance across several key drug targets. Specifically, area under the receiver operating characteristic curve (AUC) scores improved for GSK3β, DRD2, and JNK3, indicating enhanced ability to distinguish between viable and ineffective molecules. Beyond simple accuracy, the system also promotes the generation of structurally diverse compounds; metrics like IntDiv and #Circles – reaching a height (h) of 0.75 – confirm that the discovered molecules explore a wider chemical space, suggesting genuine innovation rather than reiteration of known structures and bolstering confidence in the potential for identifying truly novel therapeutics.
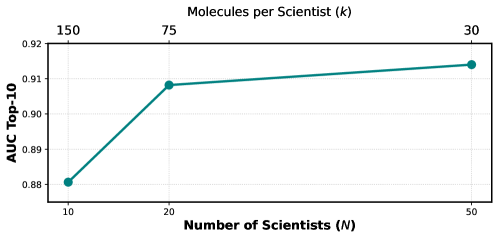
Indibator navigates the complexities of molecular discovery by prioritizing individual agent histories-a deliberate move towards manageable intricacy. The system’s reliance on publication and molecular trajectories embodies a principle of focused reasoning. As Brian Kernighan observed, “Complexity is our enemy.” Indibator doesn’t shy away from sophisticated problems, but addresses them through structured individuality, reducing overall system complexity. This approach aligns with the idea that abstractions age, principles don’t; the framework’s core tenets of fact-grounding and diverse reasoning provide lasting value in the evolving landscape of multi-agent systems.
Further Refinements
The architecture presented here, while demonstrating a capacity for differentiated reasoning within a molecular discovery context, merely addresses the symptom of homogeneity in multi-agent systems, not the underlying cause. The imposition of ‘individuality’ via publication history is, at best, a pragmatic approximation. True diversity necessitates a more fundamental exploration of agentic motivation-a synthetic analogue of intellectual curiosity, divorced from pre-existing data. The current framework remains tethered to the past; future iterations must strive for generative divergence.
A crucial limitation resides in the validation of ‘fact-grounding’. Correlation with existing literature provides a measure of consistency, not necessarily truth. Molecular space is vast, and the published record represents a biased and incomplete sampling. The system’s confidence in its assertions, therefore, warrants careful scrutiny. Objective metrics for assessing the novelty and potential utility of generated hypotheses are paramount; simply replicating known science is a computational triviality.
Ultimately, the pursuit of intelligent systems in this domain demands a shift in perspective. The goal is not to automate existing scientific processes, but to augment-and potentially surpass-human intuition. This requires embracing controlled stochasticity, accepting the inevitability of error, and prioritizing the exploration of the unexpected. Sentiment is a byproduct of structure; the elegance of a solution lies not in its complexity, but in its essential simplicity.
Original article: https://arxiv.org/pdf/2602.01815.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Clash of Clans Unleash the Duke Community Event for March 2026: Details, How to Progress, Rewards and more
- eFootball 2026 Jürgen Klopp Manager Guide: Best formations, instructions, and tactics
- MLBB x KOF Encore 2026: List of bingo patterns
- Brawl Stars February 2026 Brawl Talk: 100th Brawler, New Game Modes, Buffies, Trophy System, Skins, and more
- Gold Rate Forecast
- eFootball 2026 Starter Set Gabriel Batistuta pack review
- Magic Chess: Go Go Season 5 introduces new GOGO MOBA and Go Go Plaza modes, a cooking mini-game, synergies, and more
- Overwatch Domina counters
- ‘The Mandalorian and Grogu’ Trailer Finally Shows What the Movie Is Selling — But is Anyone Buying?
- Brent Oil Forecast
2026-02-04 04:15