Author: Denis Avetisyan
A new platform, ECGomics, is streamlining the discovery of digital biomarkers from electrocardiograms to improve cardiovascular health assessment.

ECGomics integrates expert-defined features with deep learning for multidimensional biomarker extraction, enhancing diagnostic accuracy and data efficiency.
Conventional electrocardiogram (ECG) analysis often faces a trade-off between interpretable, expert-driven features and the high accuracy but limited transparency of deep learning approaches. To address this, we present ‘ECGomics: An Open Platform for AI-ECG Digital Biomarker Discovery’, a novel framework that systematically deconstructs cardiac signals across multiple dimensions to generate robust, multidimensional digital biomarkers. This approach bridges the gap between handcrafted features and data-driven embeddings, yielding improved diagnostic precision and data efficiency. Will this paradigm shift enable a new era of personalized cardiovascular medicine through scalable, interpretable AI?
Beyond Simplistic Cardiac Signals: A Systems-Level Perspective
Conventional electrocardiogram (ECG) analysis, while a cornerstone of cardiac diagnostics, inherently simplifies the intricate electrical activity of the heart. This established methodology primarily focuses on identifying pre-defined waveform features – such as the P wave, QRS complex, and T wave – as interpreted by clinicians. However, cardiac signals exhibit substantial inter-individual variability and are often influenced by subtle physiological nuances that fall outside these standardized parameters. Consequently, critical information regarding early-stage cardiac dysfunction or atypical presentations can be overlooked. The reliance on expert-defined features introduces a degree of subjectivity and limits the capacity to detect complex patterns indicative of underlying pathology, necessitating more comprehensive analytical approaches capable of capturing the full breadth of cardiac electrical behavior.
The shift towards preventative healthcare is driving a substantial need for continuous and detailed cardiac monitoring, extending far beyond infrequent clinical check-ups. This demand isn’t simply for more data, but for meaningful data; traditional ECG analysis, while valuable, often captures only a snapshot in time and relies on pre-defined features that may miss subtle indicators of developing cardiac issues. Consequently, research is increasingly focused on methodologies capable of extracting a more complete picture from ECG signals-analyzing waveform morphology, intervals, and variations with greater precision to identify early warning signs of arrhythmias, ischemia, or structural heart disease. These advanced techniques aim to provide a more nuanced understanding of cardiac function, facilitating timely interventions and ultimately improving patient outcomes through proactive, rather than reactive, cardiac care.
Current cardiac analysis techniques often dissect electrocardiogram (ECG) signals into isolated characteristics – waveform durations, amplitude variations, and frequency components – yet struggle to synthesize these diverse features into a unified clinical picture. This fragmented approach limits the ability to detect subtle, complex patterns indicative of early-stage cardiac dysfunction or predict adverse events. The challenge lies not simply in identifying individual signal anomalies, but in understanding how these characteristics interact and contribute to the overall cardiac state. Consequently, clinicians often face difficulty translating raw signal data into actionable insights, hindering proactive risk assessment and personalized treatment strategies. A truly effective paradigm requires an integrative framework capable of contextualizing disparate signal features and delivering a cohesive, clinically relevant interpretation.
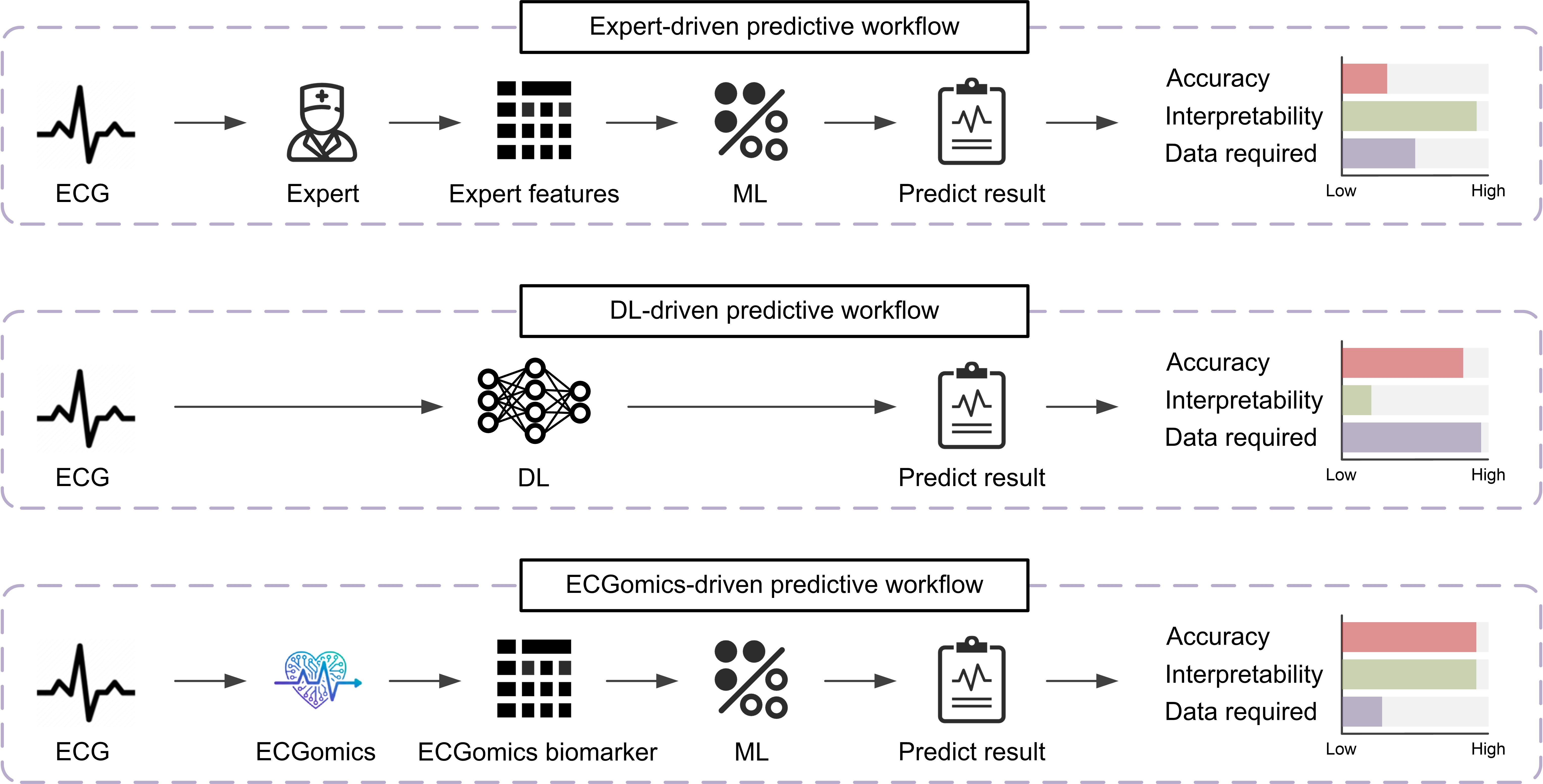
Deconstructing the Cardiac Signal: A Holistic ECGomics Approach
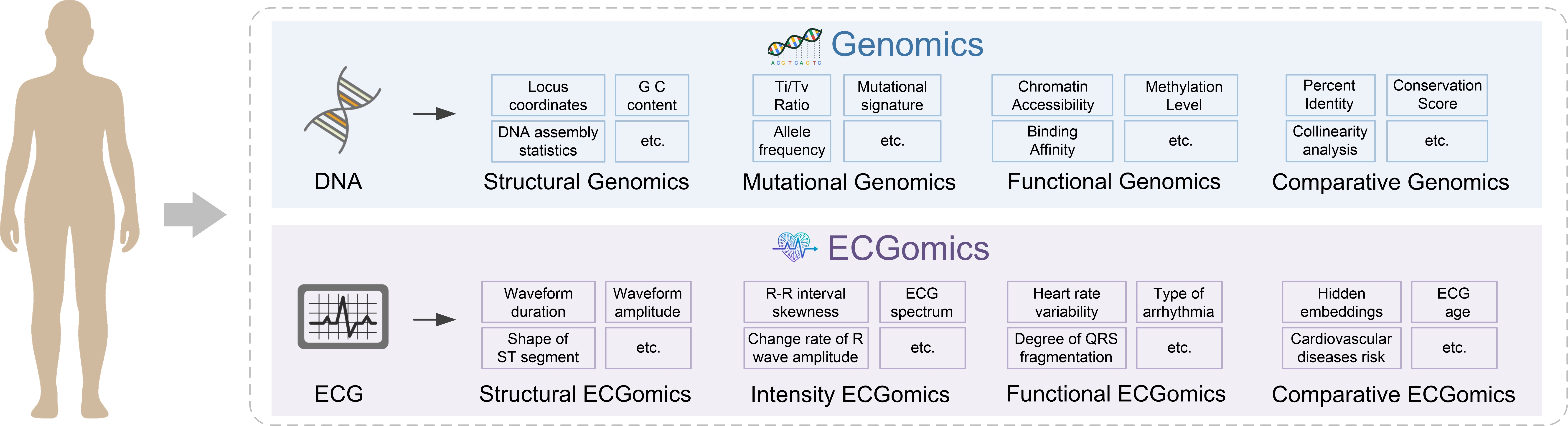
ECGomics utilizes a multidimensional analytical framework to move beyond traditional single-parameter electrocardiographic analysis. This framework deconstructs cardiac signals into three core dimensions: structural, which examines waveform morphology and timing intervals like QRS duration and PR interval; functional, focusing on dynamic changes within the signal, such as heart rate variability and repolarization characteristics; and intensity, quantifying the amplitude of various waveform components – P, QRS, and T waves. By analyzing these dimensions in concert, ECGomics provides a more holistic and detailed characterization of cardiac electrophysiology than conventional methods, allowing for the identification of subtle patterns and biomarkers indicative of underlying cardiac conditions.
ECGomics utilizes deep learning techniques, specifically employing the Net1D architecture, to surpass the feature extraction capabilities of conventional electrocardiogram (ECG) analysis. Traditional methods typically rely on a limited set of hand-engineered features, such as QRS duration and amplitude. In contrast, Net1D, a one-dimensional convolutional neural network, automatically learns complex patterns directly from the raw ECG signal. This is further augmented by FeatureDB, a curated repository of learned features, allowing ECGomics to identify a significantly broader spectrum of morphological and dynamic characteristics. The increased feature dimensionality enables a more granular and comprehensive assessment of cardiac function and pathology than is achievable with standard ECG interpretation.
Applying the principles of ‘Omics research – traditionally used in genomics, proteomics, and metabolomics – to cardiac signals involves a holistic, systems-level analysis of heart health. Rather than focusing on isolated parameters, this ECGomics approach treats the electrocardiogram as a complex dataset representing multiple interacting physiological processes. This allows for the identification of patterns and biomarkers indicative of cardiac dysfunction that might be missed by traditional single-feature analysis. By quantifying a broad spectrum of signal characteristics and their interrelationships, ECGomics aims to move beyond descriptive diagnostics toward a predictive and personalized understanding of cardiovascular disease, similar to the comprehensive insights gained from other ‘Omics fields.
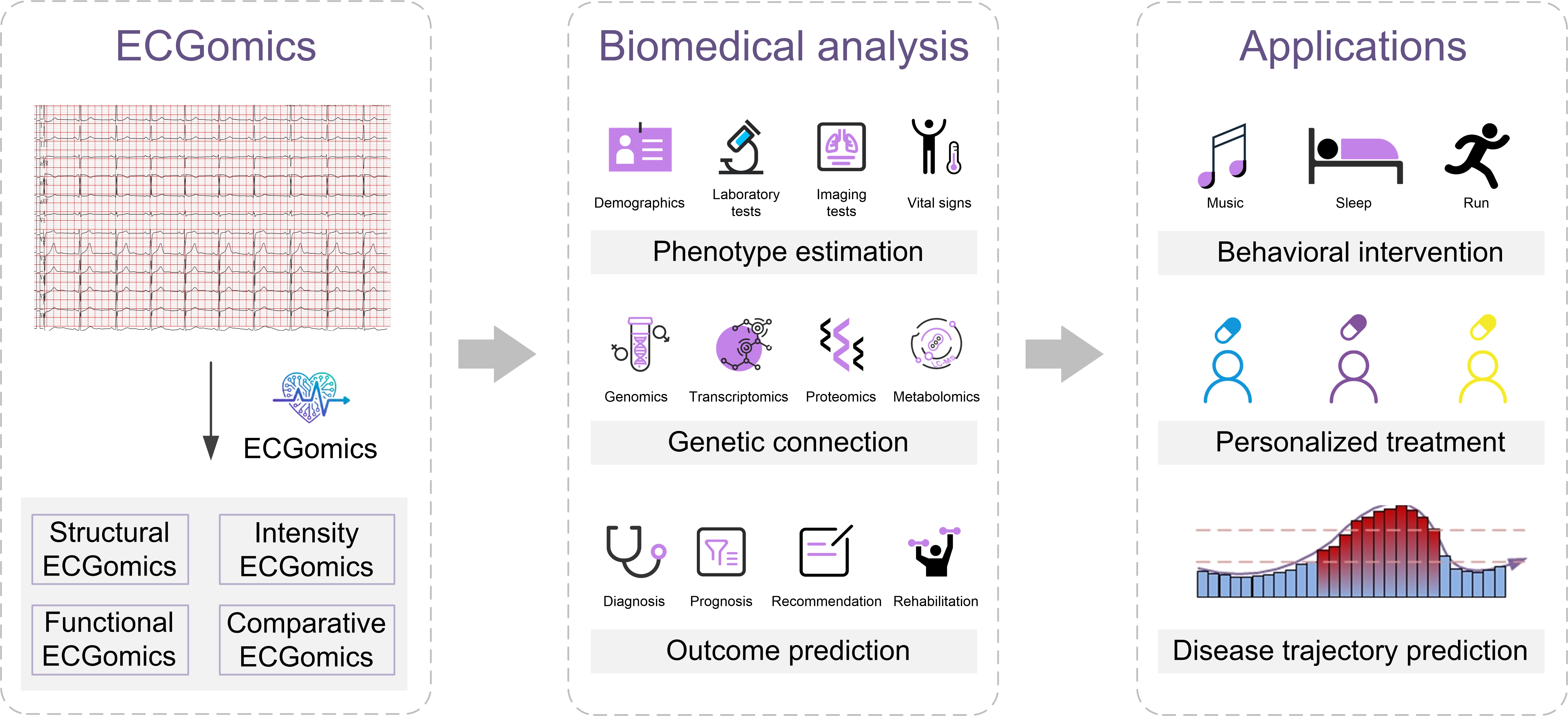
Unlocking Clinical Insights: Digital Biomarkers Derived from ECGomics
ECGomics leverages advanced analysis of electrocardiogram (ECG) data to identify digital biomarkers indicative of conditions beyond cardiac arrhythmias. While traditional ECG interpretation primarily focuses on identifying irregular heartbeats, ECGomics examines subtle waveform characteristics and intervals to assess the presence of coronary stenosis. This approach analyzes complex patterns within the ECG signal that may not be readily apparent through conventional methods, allowing for the detection of ischemic heart disease indicators. Studies demonstrate ECGomics achieves a sensitivity of up to 84.8% in coronary stenosis detection, exceeding the performance of standard ECG interpretation in identifying this condition.
XGBoost, a gradient boosting algorithm, is employed to enhance the predictive capability of digital biomarkers for cardiac events. Specifically, implementation of XGBoost in analyzing ECG-derived biomarkers achieved an Area Under the Curve (AUC) of 0.872 in predicting atrial fibrillation recurrence. This indicates a substantial improvement in predictive accuracy compared to traditional methods, allowing for earlier and more reliable identification of patients at risk of recurrent atrial fibrillation. The algorithm functions by iteratively refining predictions based on weighted combinations of features extracted from the ECG signal, optimizing for performance on established datasets.
Comparative ECGomics leverages foundation models, specifically ECGFounder and CardioLearn, to analyze individual electrocardiogram (ECG) signals in relation to large population datasets. This benchmarking process allows for the identification of subtle anomalies indicative of cardiac conditions. Studies demonstrate that ECGomics achieves a sensitivity of up to 84.8% in detecting coronary stenosis, representing an improvement over standard ECG interpretation. The diagnostic accuracy is further quantified by an Area Under the Curve (AUC) of 0.847 for stenosis detection, indicating a robust ability to differentiate between patients with and without the condition.
The Future of Cardiac Monitoring: From Reactive to Proactive and Personalized
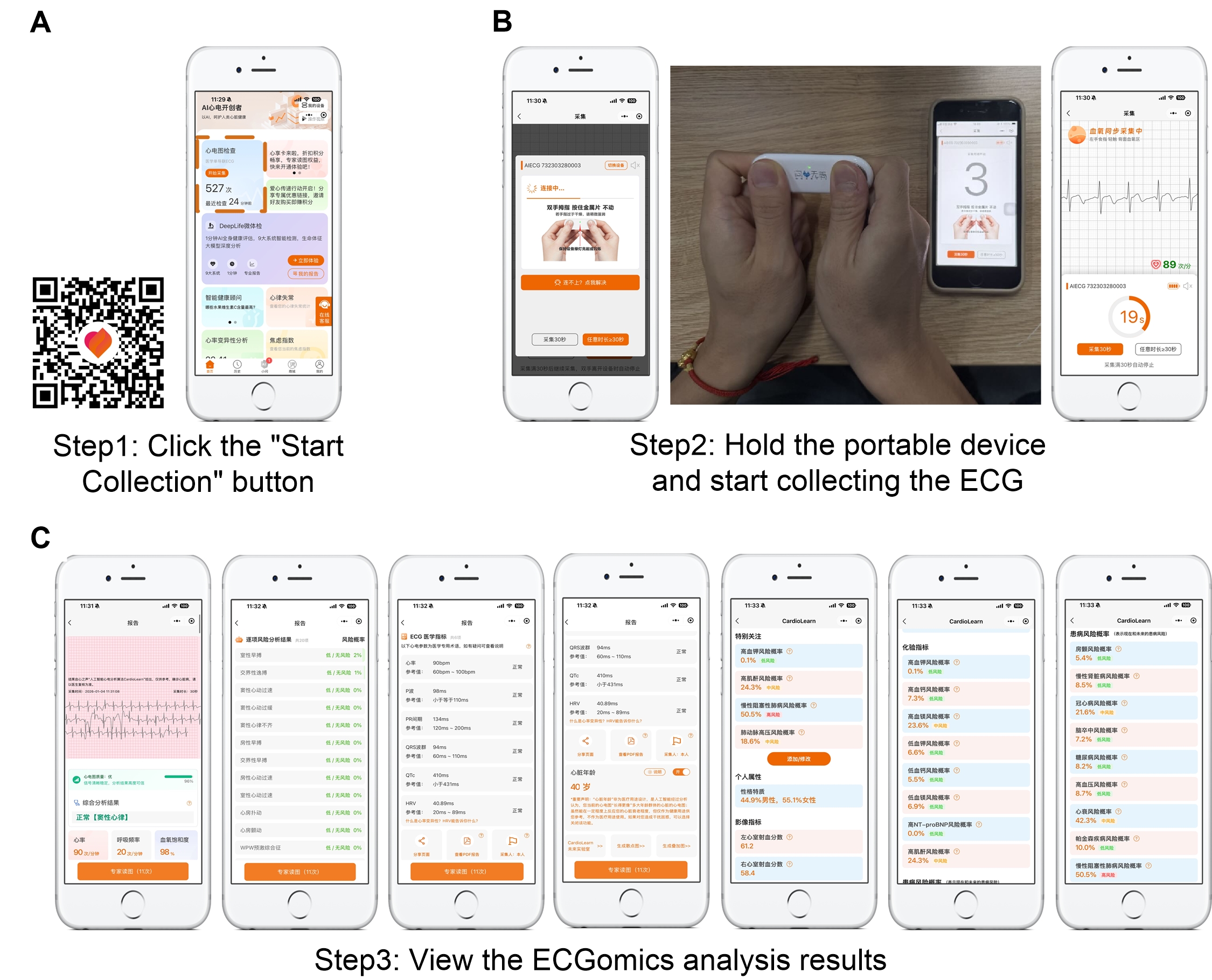
The advent of portable electrocardiogram (ECG) devices, when paired with advanced ECGomics analysis, is fundamentally shifting cardiac monitoring from reactive hospital-based assessments to proactive, continuous surveillance in everyday life. These compact devices allow for the uninterrupted capture of a patient’s cardiac rhythm and electrical activity, transmitting data for real-time interpretation using sophisticated algorithms. ECGomics, a data-driven approach, moves beyond simple arrhythmia detection, extracting a wealth of information from the ECG signal to identify subtle indicators of cardiac stress or dysfunction often missed in traditional check-ups. This continuous stream of physiological data empowers clinicians to detect anomalies early, potentially preventing acute events like heart attacks or strokes, and facilitates a deeper understanding of an individual’s cardiac health trajectory outside the limitations of a clinical visit.
Continuous cardiac monitoring, facilitated by portable ECG devices, moves beyond reactive diagnosis to proactive health management by identifying subtle deviations in heart function that might otherwise go unnoticed. These changes, often preceding acute cardiac events, represent critical opportunities for intervention. By tracking parameters like heart rate variability and QT intervals over extended periods, clinicians can establish personalized baselines and detect even minor shifts indicative of developing problems. This early warning system allows for timely adjustments to medication, lifestyle recommendations, or further diagnostic testing, potentially averting serious complications such as heart attacks, strokes, or sudden cardiac arrest – effectively shifting the paradigm from treating illness to preventing it.
The advent of digital biomarkers is poised to revolutionize cardiac care by enabling highly personalized treatment approaches. Recent studies demonstrate the robust diagnostic accuracy of these tools, consistently exceeding 0.900 in validation tests and showing strong agreement with established clinical benchmarks – notably, a correlation of 0.957 for heart rate and 0.774 for QT intervals when applied to maternal health monitoring. Furthermore, ECGomics-based analysis demonstrates a remarkable ability to identify cardiac irregularities; achieving 84.2% sensitivity and 97.5% specificity in detecting arrhythmias during maternal health monitoring. This level of precision promises to move beyond reactive treatment towards proactive, individualized interventions, optimizing patient outcomes and potentially preventing critical cardiac events before they occur.
The presented ECGomics framework embodies a systemic approach to cardiovascular assessment, mirroring the interconnectedness of urban infrastructure. Just as a city’s functionality relies on the seamless interaction of its components, ECGomics integrates expert-defined features with deep learning, creating a holistic diagnostic tool. This echoes the principle that structure dictates behavior; the framework’s design-its integration of varied data streams-directly influences its capacity to yield accurate and interpretable digital biomarkers. Friedrich Nietzsche observed, “There are no facts, only interpretations.” ECGomics doesn’t merely present data; it provides a structured means of interpreting electrocardiograms, moving beyond raw signals to meaningful clinical insights. The system’s evolution, without wholesale reconstruction, facilitates continuous improvement and adaptation to new data and clinical needs.
Where the Signal Leads
The introduction of ECGomics represents, predictably, not an arrival, but a re-centering. The pursuit of digital biomarkers from electrocardiography has, for some time, been bifurcated – a struggle between the elegance of end-to-end deep learning and the stubborn insistence of feature engineering. This framework attempts to synthesize those approaches, but the true test will lie in demonstrating how this integration alters the systemic behavior of diagnostic models. Simply achieving higher accuracy is insufficient; the system’s vulnerabilities, its points of failure when confronted with novel pathologies or population shifts, must be mapped with equal rigor.
A crucial next step involves expanding the scope of inquiry. ECGomics, as presented, focuses on the signal itself. However, the cardiovascular system is, of course, inextricably linked to other physiological processes. The framework’s architecture invites, almost demands, integration with multi-modal data – genomic information, proteomic profiles, lifestyle metrics. Such expansion, while computationally demanding, is inevitable, and will reveal whether the extracted biomarkers represent fundamental properties of disease or merely correlations shaped by confounding variables.
Finally, the question of interpretability remains paramount. While feature engineering provides a degree of transparency, the deep learning components still operate as, to some extent, ‘black boxes’. The challenge is not merely to identify biomarkers, but to understand how those biomarkers reflect underlying physiological mechanisms. Until that understanding is achieved, the system will remain a sophisticated tool for prediction, but not a pathway to true understanding.
Original article: https://arxiv.org/pdf/2601.15326.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Gold Rate Forecast
- Star Wars Fans Should Have “Total Faith” In Tradition-Breaking 2027 Movie, Says Star
- Christopher Nolan’s Highest-Grossing Movies, Ranked by Box Office Earnings
- Jessie Buckley unveils new blonde bombshell look for latest shoot with W Magazine as she reveals Hamnet role has made her ‘braver’
- KAS PREDICTION. KAS cryptocurrency
- Country star Thomas Rhett welcomes FIFTH child with wife Lauren and reveals newborn’s VERY unique name
- eFootball 2026 is bringing the v5.3.1 update: What to expect and what’s coming
- eFootball 2026 Jürgen Klopp Manager Guide: Best formations, instructions, and tactics
- Are Halstead & Upton Back Together After The 2026 One Chicago Corssover? Jay & Hailey’s Future Explained
- Genshin Impact Version 6.5 Leaks: List of Upcoming banners, Maps, Endgame updates and more
2026-01-24 03:52