Author: Denis Avetisyan
Researchers are shifting the focus from simply predicting drug-drug interactions to building a more generalizable understanding of how molecules interact with each other.
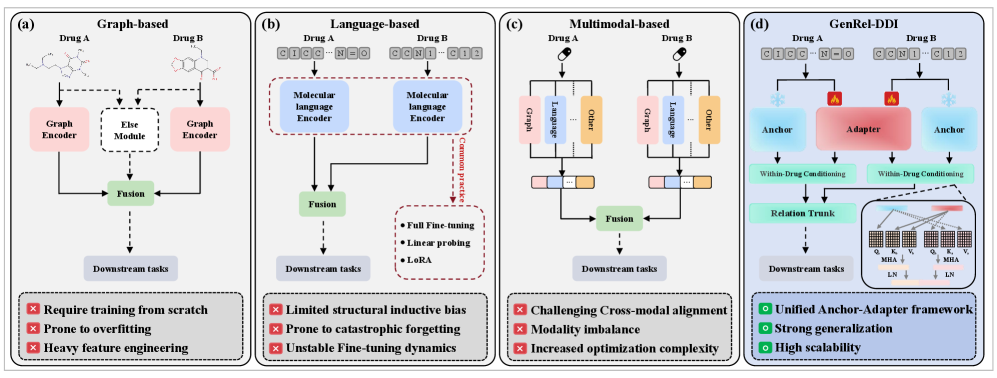
This review introduces GenRel-DDI, a novel approach that reframes drug-drug interaction prediction as a relation learning problem, achieving state-of-the-art performance through decoupled molecular representations and cross-attention mechanisms.
Despite advances in computational prediction, current drug-drug interaction (DDI) models often struggle to generalize to realistic scenarios involving novel drug combinations. This limitation motivates ‘Rethinking Drug-Drug Interaction Modeling as Generalizable Relation Learning’, which proposes a novel framework, GenRel-DDI, that reframes DDI prediction as a relation-centric learning problem-decoupling molecular identities from interaction patterns to improve transferability. Through extensive benchmarking, particularly on strict entity-disjoint evaluations, GenRel-DDI demonstrates state-of-the-art performance and highlights the efficacy of relation learning for robust DDI prediction. Could this relation-centric approach unlock more reliable and scalable strategies for identifying potentially adverse drug interactions in clinical settings?
The Challenge of Polypharmacy: A Need for Predictive Precision
The increasing prevalence of polypharmacy – the simultaneous use of multiple medications – presents a growing challenge to patient safety, demanding robust methods for predicting drug interactions. As individuals age and manage multiple chronic conditions, the likelihood of receiving combinations of drugs not thoroughly tested for synergistic or antagonistic effects rises significantly. These drug-drug interactions can lead to reduced medication efficacy, unexpected side effects, and even life-threatening complications. Consequently, the ability to proactively identify potential adverse interactions becomes paramount, necessitating a shift towards predictive models that can reliably assess the safety of increasingly complex medication regimens and safeguard patient well-being.
Current drug interaction prediction methods often falter when faced with novel drug combinations not previously documented in training datasets. These techniques, frequently reliant on identifying patterns from known pairings, demonstrate limited ability to extrapolate to unseen scenarios, creating a substantial risk for patients undergoing polypharmacy. This generalization challenge stems from the inherent complexity of biological systems; interactions aren’t simply additive but can be synergistic or antagonistic, influenced by individual patient factors like genetics and metabolism. Consequently, a drug combination deemed safe based on existing data may unexpectedly trigger adverse effects, highlighting the urgent need for more robust and adaptable predictive models capable of navigating the vast, unexplored space of potential drug interactions and safeguarding patient health.
Predicting how drugs will interact in vivo presents a substantial challenge, as the sheer complexity of biological systems necessitates methods that transcend traditional approaches. Current predictive models often falter when faced with novel drug combinations-those not previously documented or studied-highlighting their limited ability to generalize beyond known interactions. This limitation stems from the fact that drug effects aren’t simply additive; they arise from intricate, multi-faceted relationships influenced by individual patient factors, metabolic pathways, and even epigenetic variations. Consequently, research is increasingly focused on developing innovative computational strategies-including machine learning and network pharmacology-designed to infer interactions based on drug structure, target proteins, and broader biological context, rather than relying solely on pre-existing data. These advanced techniques aim to move beyond simple pattern recognition and towards a more holistic understanding of drug behavior, ultimately improving patient safety in an era of increasingly complex polypharmacy.
Reframing DDI Prediction: Focusing on Relationships
GenRel-DDI addresses limitations in traditional drug-drug interaction (DDI) prediction models which are heavily reliant on specific drug features, hindering performance with novel compounds. This framework decouples drug identity from the interaction prediction process by focusing on the relational aspects of interactions rather than intrinsic drug properties. This is achieved through a system designed to generalize to previously unseen drugs without requiring retraining on their specific characteristics, thereby improving predictive capability for emerging pharmaceutical compounds and addressing the cold-start problem common in DDI prediction tasks. The core innovation lies in the ability to predict interactions based on the relationships between drugs, independent of their individual identities.
Relation learning, as applied to drug-drug interaction (DDI) prediction, shifts the focus from encoding specific drug characteristics to modeling the inherent relationships between drugs that govern interaction potential. Traditional methods often rely on feature vectors representing individual drugs, limiting generalization to novel compounds. By instead learning the relational structure – how drugs interact based on their chemical properties and biological targets – the model can infer interactions for previously unseen drugs without requiring their explicit feature representation. This approach aims to capture the underlying principles of interaction, such as specific functional groups causing reactivity or shared target proteins, rather than memorizing patterns associated with known drug pairs. Consequently, the model’s predictive capability is less dependent on the specific attributes of individual drugs and more reliant on the learned relational principles.
The Anchor-Adapter Framework utilizes a two-part architecture for drug-drug interaction (DDI) prediction. A fixed, pretrained encoder serves as the “anchor,” processing input drug representations and establishing a foundational feature space. Complementing this is a trainable “relation module,” or “adapter,” which takes the anchor’s output and learns to model the specific relationship between drug pairs relevant to interaction prediction. This decoupling allows the pretrained encoder to generalize across diverse drug compounds, while the adapter focuses on learning interaction-specific patterns, improving performance on both known and novel drugs. The adapter’s parameters are updated during training, while the anchor remains fixed, reducing computational cost and mitigating overfitting.
Encoding Drug Representations: Building a Foundation for Prediction
GenRel-DDI employs pretrained transformer-based encoders to convert molecular structures into numerical representations, or embeddings. Specifically, the model leverages architectures including MolT5, MoLFormer, and ChemBERTa-3, each trained on extensive chemical datasets to learn meaningful features of molecular compounds. These encoders output fixed-length vectors that capture the chemical properties and structural information of each drug, providing a standardized input for downstream interaction prediction tasks. The use of pretrained models mitigates the need for extensive training data and allows for the transfer of knowledge learned from large-scale chemical corpora, resulting in more robust and generalizable molecular representations.
Drug representations generated by pretrained encoders are combined, or fused, within the GenRel-DDI framework to facilitate prediction of drug-drug interactions. This fusion process allows the model to analyze the combined characteristics of multiple molecules simultaneously, rather than treating them in isolation. By integrating these encoded features, the system can identify complex relationships – including synergistic or antagonistic effects – that might not be apparent from individual molecular properties alone. The resulting interaction prediction benefits from a holistic understanding of chemical interplay, leading to improved accuracy in forecasting potential drug interactions.
The Molecule-Centric Pipeline in GenRel-DDI establishes a standardized process for converting molecular structures into numerical representations suitable for machine learning. This pipeline initially focuses on individual drug encoding, creating embeddings that capture intrinsic chemical properties before considering interactions. This foundational step is crucial as it allows the model to learn drug characteristics independent of specific interaction contexts, improving generalization and enabling the prediction of novel drug-drug interactions. The resulting embeddings serve as input features for downstream interaction analysis, providing a consistent and informative basis for assessing relationships between drugs.
![Performance on datasets S1-S3, measured by AUPR, demonstrates that MeTDDI achieves competitive results with both frozen ([latex] \ast [/latex]) and fully fine-tuned pretrained encoders, exhibiting consistent performance across five independent runs (mean ± std).](https://arxiv.org/html/2601.15771v1/x4.png)
Rigorous Evaluation: Demonstrating Generalization and Performance
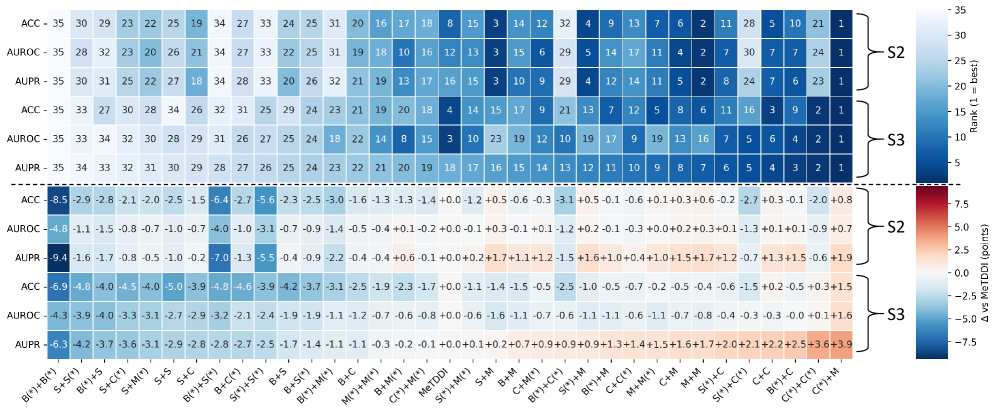
Evaluation of the GenRel-DDI framework utilizes established benchmark datasets, specifically MeTDDI and DDInter, to quantify performance. To ensure reliable assessment and mitigate inflated results stemming from data leakage, overlap-controlled splits are implemented during the evaluation process. These splits are designed to prevent information from the training set influencing the model’s performance on the test set by carefully managing the presence of shared entities and interactions between the two. This rigorous methodology facilitates a more accurate and trustworthy evaluation of the framework’s ability to generalize to unseen data.
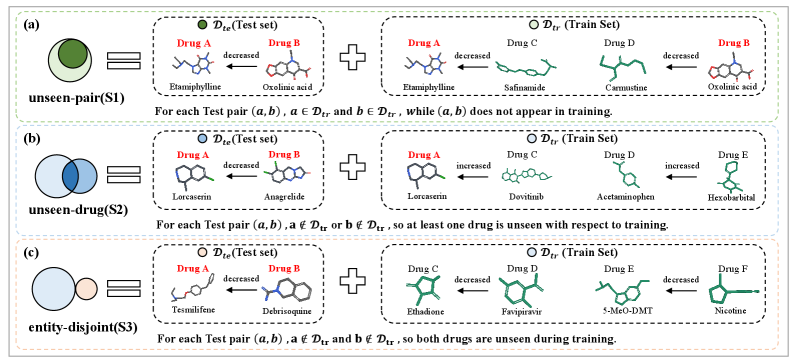
Generalization performance was assessed using specialized evaluation splits designed to isolate specific challenges. The Unseen-Pair split evaluates the model’s ability to predict interactions for drug and target combinations not present in the training data. The Unseen-Drug split tests performance on drugs entirely absent from the training set, while the Entity-Disjoint split assesses the model’s capability to generalize to entirely new entities – both drugs and targets – not encountered during training. These splits provide a more robust evaluation than standard train/test splits by specifically targeting the model’s ability to extrapolate beyond known data, thus measuring true generalization capability.
GenRel-DDI achieved an Area Under the Receiver Operating Characteristic curve (AUROC) of 99.3% when evaluated on the MeTDDI S1 dataset. This score indicates a very high ability to distinguish between known drug-drug interactions and non-interactions within this benchmark. The MeTDDI S1 dataset is a standard resource for evaluating DDI prediction models, and an AUROC of 99.3% represents a substantial level of predictive accuracy, demonstrating the framework’s capacity to correctly identify true interactions while minimizing false positives in this specific evaluation setting.
GenRel-DDI demonstrates a 3.9% improvement in Area Under the Precision-Recall curve (AUPR) on the S3 split of the MeTDDI dataset when compared to the baseline model. This performance gain is particularly significant as the S3 split represents a more challenging evaluation scenario, designed to assess the model’s capability to identify interactions under conditions of increased complexity or data scarcity. The AUPR metric quantifies the precision of positive predictions at varying recall levels, making it a robust indicator of the model’s ability to accurately predict drug-drug interactions even when faced with imbalanced datasets or subtle interaction patterns.
GenRel-DDI establishes a new state-of-the-art performance level across multiple established drug-drug interaction benchmarks. Specifically, the framework achieves the highest reported Area Under the Receiver Operating Characteristic curve (AUROC) and Area Under the Precision-Recall curve (AUPR) scores on the DeepDDI, ZhangDDI, and ChChDDI datasets. This performance surpasses previously published results, indicating an improved capacity to accurately predict drug-drug interactions as measured by these standardized evaluation metrics and datasets.
The pursuit of increasingly intricate models often obscures a fundamental truth: simplicity yields strength. This work, with its decoupling of molecular representations and interaction patterns in GenRel-DDI, embodies that principle. They called it a framework to hide the panic, perhaps, but it is, in essence, a recognition that generalization isn’t achieved through complexity, but through elegant abstraction. As Grace Hopper observed, “It’s easier to ask forgiveness than it is to get permission.” This sentiment applies perfectly; the researchers didn’t seek permission to dismantle conventional approaches, they simply forged ahead with a more direct, and ultimately more effective, path toward robust drug-drug interaction prediction, even in challenging entity-disjoint splits.
What’s Next?
The decoupling of molecular encoding from interaction modeling, as demonstrated, offers a reprieve from the tendency to overcomplicate. GenRel-DDI’s performance in entity-disjoint splits suggests a move toward genuine generalization, though the simplicity should not be mistaken for completion. The framework presently addresses prediction; the question of why interactions occur remains largely untouched. Future work must resist the urge to simply add layers of complexity-more parameters are not, inherently, more understanding.
A critical next step involves extending this relation-learning approach beyond pairwise interactions. Biological systems are rarely so neatly organized. Modeling multi-drug combinations, or the interplay between drugs and proteins, will demand a refinement of the current methodology. The current reliance on cross-attention, while effective, may prove insufficient for representing the intricate dependencies inherent in complex biological networks.
Ultimately, the pursuit of perfect predictive accuracy risks obscuring a more fundamental goal: a concise and interpretable model of drug action. The elegance of GenRel-DDI lies in its restraint. The field would benefit from further exploration of methods that prioritize clarity and parsimony, recognizing that the most powerful models are often those that reveal the essential structure underlying the noise.
Original article: https://arxiv.org/pdf/2601.15771.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Gold Rate Forecast
- Star Wars Fans Should Have “Total Faith” In Tradition-Breaking 2027 Movie, Says Star
- Christopher Nolan’s Highest-Grossing Movies, Ranked by Box Office Earnings
- KAS PREDICTION. KAS cryptocurrency
- Jessie Buckley unveils new blonde bombshell look for latest shoot with W Magazine as she reveals Hamnet role has made her ‘braver’
- Country star Thomas Rhett welcomes FIFTH child with wife Lauren and reveals newborn’s VERY unique name
- eFootball 2026 is bringing the v5.3.1 update: What to expect and what’s coming
- eFootball 2026 Jürgen Klopp Manager Guide: Best formations, instructions, and tactics
- Clash of Clans Unleash the Duke Community Event for March 2026: Details, How to Progress, Rewards and more
- Genshin Impact Version 6.5 Leaks: List of Upcoming banners, Maps, Endgame updates and more
2026-01-25 05:05