Author: Denis Avetisyan
A new approach harnesses schema constraints and provenance tracking to automatically extract structured data from scientific PDFs, streamlining biomedical research.
This review details a robust system for schema-constrained AI extraction, OCR, and provenance tracking to improve the reliability and scalability of biomedical evidence synthesis from full-text documents.
Despite the increasing volume of biomedical literature, extracting reliable, structured data from full-text PDFs remains a significant bottleneck for evidence synthesis. This work, ‘From Chaos to Clarity: Schema-Constrained AI for Auditable Biomedical Evidence Extraction from Full-Text PDFs’, introduces a novel system that leverages schema constraints and provenance tracking to transform complex scientific documents into analysis-ready records. By explicitly guiding model inference and ensuring deterministic merging of extracted data, the pipeline achieves both scalability and auditability-critical for high-stakes biomedical research. Could this approach unlock a new era of automated, transparent, and reliable evidence synthesis, accelerating the translation of research findings into clinical practice?
The Extraction Bottleneck: Navigating the Deluge of Scientific Literature
The exponential growth of scientific research has created a paradoxical situation: while knowledge production surges, access to that knowledge remains surprisingly limited. A significant portion of published findings resides within unstructured PDF documents – essentially digital pages of text and figures – making automated analysis exceptionally difficult. This creates a substantial bottleneck, hindering meta-analysis, systematic reviews, and the development of new hypotheses. Researchers spend countless hours manually extracting data, a process that is both time-consuming and prone to error. The inability to efficiently synthesize existing knowledge not only slows down the pace of discovery but also duplicates effort, diverting resources from genuinely novel investigations and impeding scientific progress across all disciplines.
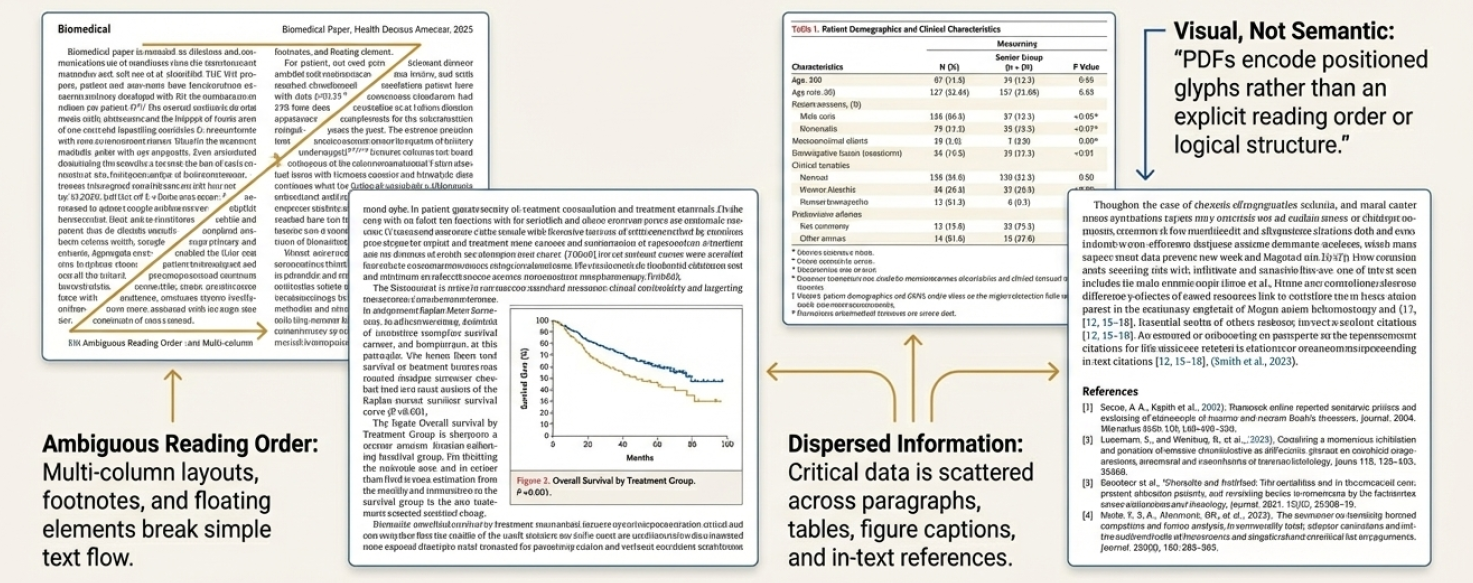
Conventional information extraction techniques, designed for structured data, frequently falter when applied to the nuanced landscape of scientific literature. These methods often rely on rigid templates and predefined patterns, proving inadequate for the variability inherent in research papers – differing section headings, inconsistent data presentation, and the pervasive use of complex tables and figures. The inherent complexity arises not only from the diverse range of scientific disciplines-each with its own reporting conventions-but also from the evolving nature of research itself. This poses a significant challenge, as accurately pinpointing key findings, experimental parameters, or statistical results requires navigating inconsistent formatting, deciphering specialized terminology, and accounting for the unique structural characteristics of each document – tasks that demand a more adaptable and intelligent approach than traditional methods can consistently deliver.
Scientific literature presents a unique extraction challenge due to the intricate formatting and diverse layouts inherent in research papers. Unlike standardized documents, PDFs often contain multi-column text, tables with varying structures, complex mathematical equations represented with [latex]\sum_{i=1}^{n} x_i[/latex], and figures embedded within the text. Accurately pinpointing key data – such as experimental parameters, statistical results, or specific chemical compounds – requires algorithms capable of deciphering these complex visual cues and contextual relationships. Traditional optical character recognition (OCR) and rule-based methods frequently falter when faced with variations in font, size, and positioning, leading to inaccurate or incomplete data extraction. Consequently, unlocking the vast potential of scientific knowledge hinges on developing sophisticated techniques that can robustly navigate these complex document structures and reliably identify the crucial information embedded within.
Document Understanding: From Pixels to Structure
Scientific PDF extraction necessitates both Optical Character Recognition (OCR) and Layout Analysis due to the complex structure of these documents. OCR converts images of text into machine-readable text, but struggles with formatting and context without further processing. Layout Analysis, conversely, determines the organization of the document, identifying elements like headings, paragraphs, tables, figures, and their spatial relationships. Combining these two approaches allows systems to not only read the text but also understand its meaning within the document’s structure, significantly improving the accuracy and reliability of information extraction from scientific literature. Without Layout Analysis, OCR output would be a flat stream of characters, lacking the contextual information needed for meaningful interpretation.
DocFormer and LayoutLM represent a shift in document understanding by employing Transformer architectures to jointly process textual content and spatial layout. Traditionally, Optical Character Recognition (OCR) produced text independent of its location on the page. These models, however, utilize the self-attention mechanism inherent in Transformers to consider the relationships between words and their visual positioning. Specifically, visual embeddings representing the location and size of text blocks are incorporated into the Transformer’s input alongside textual embeddings. This allows the model to learn contextual relationships informed by both the content and the layout, improving performance on tasks requiring an understanding of document structure, such as key-value pair extraction and table recognition. The unified approach avoids the need for separate processing pipelines and enables end-to-end training for enhanced accuracy.
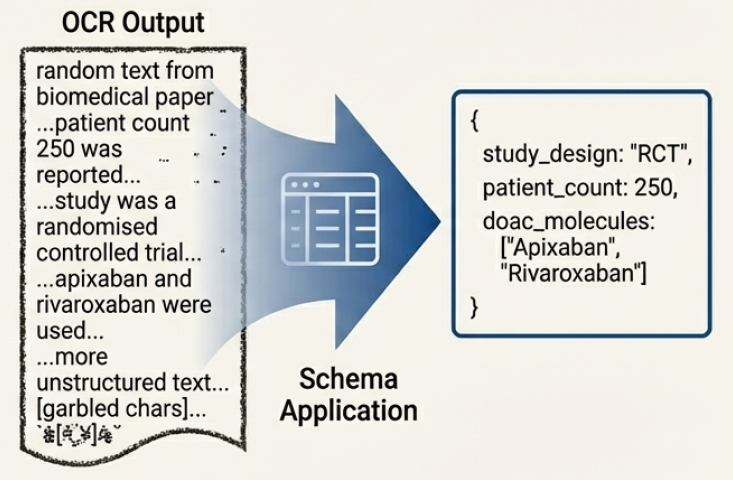
Schema-constrained extraction improves the precision of information retrieval from documents by incorporating a predefined Schema, which acts as a structural template for the expected data. This Schema defines the types of entities to be extracted – such as experimental parameters, material properties, or numerical results – and their relationships. By guiding the extraction process with this structural knowledge, the system focuses on identifying and capturing only relevant information, reducing noise and ambiguity. The Schema is typically represented as a set of labels and relationships, enabling the model to constrain its predictions and improve accuracy, particularly when dealing with complex document layouts or inconsistent formatting. This approach is especially effective in scientific and technical domains where data follows established conventions and predictable structures.
Validating Reliability: Benchmarking and Assessment
PubLayNet and DocBank are publicly available datasets designed to facilitate the development and assessment of document understanding systems. PubLayNet, comprised of over 1 million scientific articles, provides pixel-level annotations of document layout elements such as text, figures, and tables, enabling training of models for layout analysis. DocBank, a collection of over 1.2 million scientific documents, focuses on the extraction of document structure and metadata, including headers, paragraphs, and citations. These datasets offer ground truth data for supervised learning and standardized benchmarks for comparing the performance of different document understanding algorithms, specifically in areas like information extraction, document classification, and optical character recognition (OCR) post-processing.
Several software tools facilitate document information extraction. GROBID (GeneRation Of BIbliographic Data) focuses on extracting bibliographic data from scholarly PDF documents, identifying structural elements like headings, references, and affiliations. CERMINE similarly automates the extraction of text and metadata from scientific papers, emphasizing layout analysis. ParsCit specializes in citation parsing, accurately identifying and extracting citation information from text. OmniDocBench provides a standardized benchmark suite for evaluating the performance of document understanding systems across various tasks, including layout analysis, information extraction, and table detection, allowing for comparative assessment of different models and techniques.
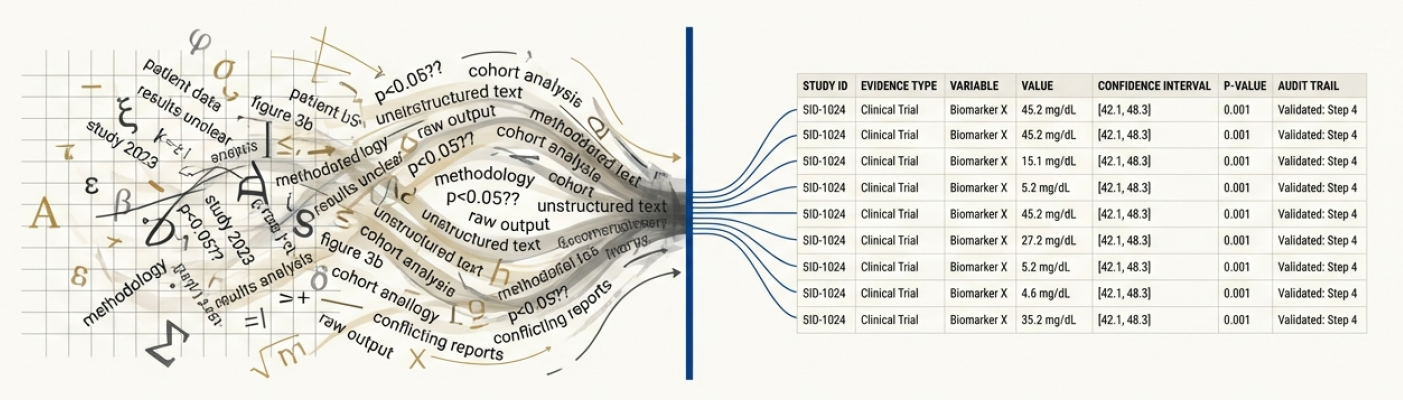
Provenance-Aware Extraction enhances data reliability by systematically recording the lineage of extracted information. This involves tracking not only the source document but also the specific algorithms, parameters, and processing steps used during extraction. Metadata associated with each extracted element includes details such as the version of the extraction tool, the date and time of processing, and any user-defined annotations or modifications. This detailed history enables verification of results, facilitates error analysis, and supports reproducibility of findings, ultimately increasing confidence in the accuracy and trustworthiness of the extracted data.
Accelerating Discovery: Synthesis and Beyond
The foundation of large-scale scientific evidence synthesis hinges on the ability to reliably extract information from the vast and growing body of published research, primarily distributed as Portable Document Format (PDF) files. Without accurate and efficient parsing of these documents, attempts to systematically review and synthesize findings are hampered by bottlenecks and potential inaccuracies. This reliance on PDF extraction isn’t merely a technical hurdle; it directly impacts the speed at which new insights can be generated, hindering progress across diverse scientific disciplines. The challenge lies not just in recognizing text, but in correctly identifying and categorizing complex scientific data – figures, tables, equations [latex]E=mc^2[/latex], and nuanced experimental details – necessitating sophisticated techniques to move beyond simple optical character recognition and towards true semantic understanding of scientific literature.
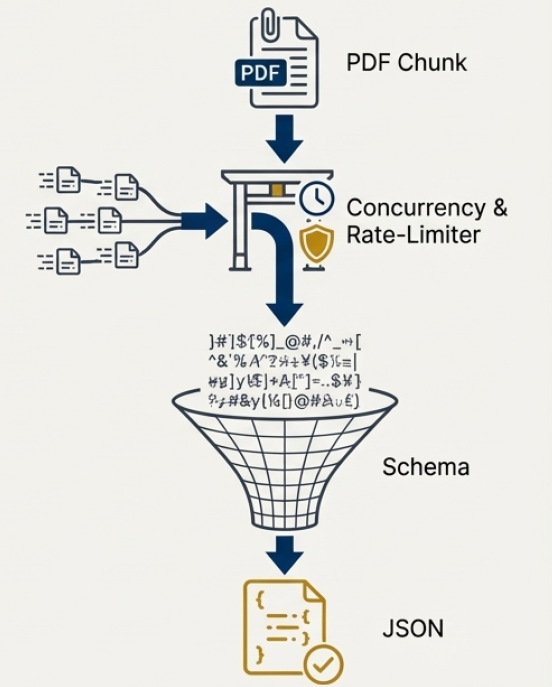
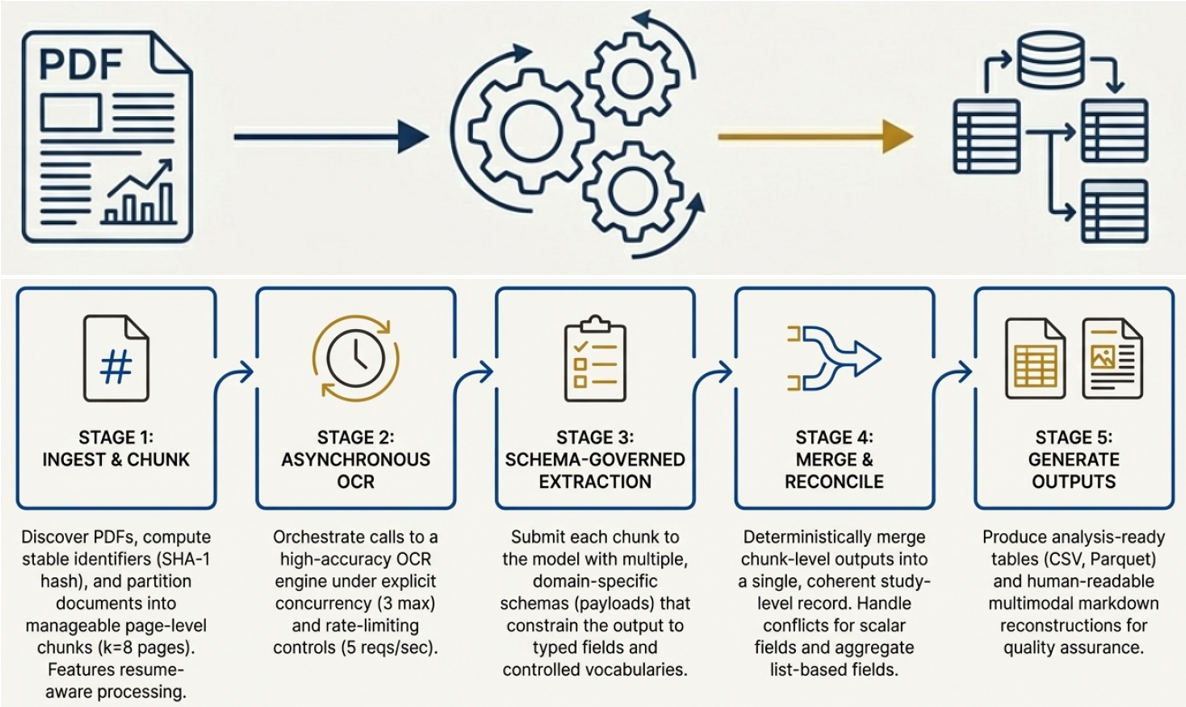
To accelerate the extraction of information from scientific PDFs, the system employs a two-pronged approach centered on page chunking and Conditional Random Fields (CRFs). Initially, each PDF is divided into smaller, manageable segments – a process known as page chunking – which allows for parallel processing and drastically reduces computational load. Subsequently, Conditional Random Fields, a type of discriminative probabilistic model, are utilized to accurately identify and categorize key elements within each chunk, such as headings, paragraphs, and figures. This machine learning technique considers the contextual relationships between elements, improving accuracy beyond simple keyword-based approaches. By combining these methods, the system achieves both high processing speed and reliable information extraction, essential for large-scale scientific evidence synthesis.
A robust evaluation of the scientific document processing pipeline involved successfully extracting data from a corpus of 734 diverse PDF articles. This large-scale test demonstrated not only the system’s functional accuracy, but also its capacity for stable throughput and scalability – crucial characteristics for handling the ever-growing volume of scientific literature. Consistent performance across the dataset confirms the system’s reliability in processing varied document structures and complexities, paving the way for its application to substantially larger collections and enabling comprehensive analyses previously hindered by processing bottlenecks. The ability to consistently process documents at scale is a key step toward automated scientific evidence synthesis.
The developed pipeline dramatically accelerates the pace of scientific data acquisition, achieving a mean document processing time of just 11 seconds. This represents a substantial improvement over traditional methods of manual extraction, which routinely require tens of minutes – and often hours – per scientific publication. Such a reduction in processing time isn’t merely incremental; it unlocks the potential for large-scale scientific evidence synthesis, enabling researchers to analyze vast quantities of literature in a timeframe previously considered impractical. This efficiency gain allows for quicker identification of key findings, accelerates meta-analysis, and ultimately facilitates more rapid advancements across diverse scientific disciplines.
The system’s capacity to process approximately 67 scientific PDFs each hour represents a substantial advancement in the speed of knowledge synthesis. This throughput, achieved through automated extraction and processing techniques, allows researchers to move beyond the limitations of manual data retrieval – a traditionally time-consuming endeavor often requiring tens of minutes per document. Consequently, large-scale analyses previously impractical due to temporal constraints become feasible, accelerating the pace of scientific discovery and enabling more comprehensive evidence-based insights across diverse fields. The ability to rapidly ingest and analyze such a volume of literature promises to reshape how research is conducted and disseminated, fostering a more dynamic and efficient scientific landscape.
The pursuit of structured data from unstructured sources, as detailed in the paper, echoes a fundamental tenet of clear thinking. It strives to distill complex information into manageable components, mirroring the reduction of noise to signal. As Bertrand Russell observed, “The point of the system is to make plain simple things.” This resonates deeply with the system’s focus on schema-constrained extraction; by imposing structure, the system minimizes ambiguity and maximizes the utility of the extracted evidence. The system’s provenance tracking, ensuring data reliability, further exemplifies this commitment to clarity, acknowledging that trust in information is paramount. The work embodies the principle that perfection isn’t about adding more complexity, but about meticulously removing unnecessary layers to reveal the essential truth within the biomedical literature.
Beyond Extraction: Towards Cognitive Documents
The presented work addresses a practical, if unglamorous, bottleneck in biomedical research. Yet, solving for reliability in data extraction only postpones a deeper question. The true challenge isn’t converting unstructured text into tables, but representing the meaning inherent in the original document – its claims, limitations, and underlying assumptions. Schema constraints, while valuable, are inherently reductive. They impose order where nuance often resides, trading information for the illusion of completeness.
Future iterations must move beyond feature engineering towards genuinely cognitive document understanding. This necessitates integrating extraction with reasoning capabilities – allowing the system not merely to identify data points, but to assess their validity and contextualize them within the broader scientific landscape. Provenance tracking, currently focused on extraction steps, should extend to capture the intellectual lineage of claims – tracing arguments back to their foundational premises.
Ultimately, the goal isn’t simply to automate evidence synthesis, but to build systems that can critically appraise evidence. This requires embracing uncertainty, acknowledging the limits of current knowledge, and designing for graceful degradation when faced with ambiguity. The pursuit of perfect data is a fool’s errand; the art lies in managing imperfection with elegance and transparency.
Original article: https://arxiv.org/pdf/2601.14267.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- VCT Pacific 2026 talks finals venues, roadshows, and local talent
- Will Victoria Beckham get the last laugh after all? Posh Spice’s solo track shoots up the charts as social media campaign to get her to number one in ‘plot twist of the year’ gains momentum amid Brooklyn fallout
- Dec Donnelly admits he only lasted a week of dry January as his ‘feral’ children drove him to a glass of wine – as Ant McPartlin shares how his New Year’s resolution is inspired by young son Wilder
- The five movies competing for an Oscar that has never been won before
- Vanessa Williams hid her sexual abuse ordeal for decades because she knew her dad ‘could not have handled it’ and only revealed she’d been molested at 10 years old after he’d died
- Binance’s Bold Gambit: SENT Soars as Crypto Meets AI Farce
- Invincible Season 4’s 1st Look Reveals Villains With Thragg & 2 More
- SEGA Football Club Champions 2026 is now live, bringing management action to Android and iOS
- How to watch and stream the record-breaking Sinners at home right now
- Jason Statham, 58, admits he’s ‘gone too far’ with some of his daring action movie stunts and has suffered injuries after making ‘mistakes’
2026-01-23 06:00