Author: Denis Avetisyan
A new study identifies urinary metabolic markers that accurately distinguish individuals with ADHD, offering a potential path toward more objective diagnosis.
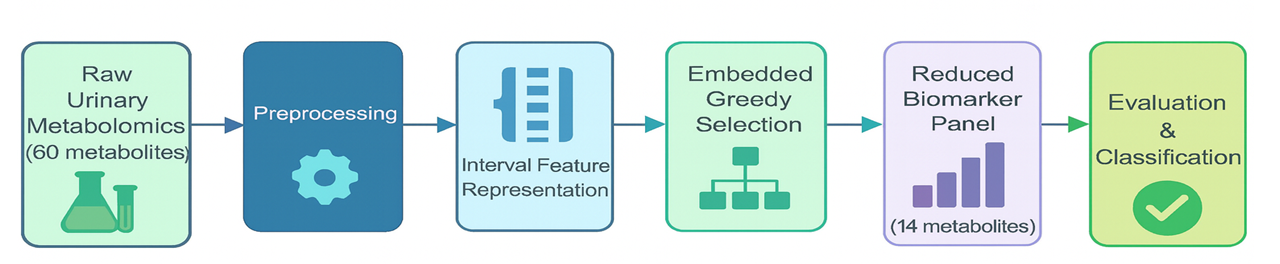
Interpretable machine learning, specifically a Closest Resemblance classifier with embedded feature selection, was used to identify key metabolites associated with ADHD.
Despite increasing prevalence, Attention Deficit Hyperactivity Disorder (ADHD) lacks robust, objective diagnostic tools, hindering precision psychiatry efforts. This challenge is addressed in ‘Metabolomic Biomarker Discovery for ADHD Diagnosis Using Interpretable Machine Learning’, which integrates urinary metabolomics with a novel machine learning approach to identify biochemical signatures of the disorder. The study demonstrates that a Closest Resemblance classifier, utilizing a reduced panel of 14 metabolites, accurately distinguishes individuals with ADHD from controls with exceptional performance and improved interpretability. Could this framework pave the way for targeted metabolomic assays and ultimately, point-of-care diagnostics for ADHD?
The Illusion of Objective Assessment
Attention-deficit/hyperactivity disorder (ADHD) diagnosis currently depends significantly on behavioral evaluations interpreted through frameworks like the DSM-5 and ICD-11. While these diagnostic manuals provide standardized criteria, their reliance on observations of behavior-often reported by parents or teachers-introduces a degree of subjectivity. This subjective element can lead to diagnostic inaccuracies, with symptoms potentially misattributed to other factors or minimized due to differing interpretations. Consequently, individuals who genuinely require intervention may face delayed diagnosis and access to appropriate support, hindering academic, social, and occupational development. The inherent limitations of behavioral assessments highlight the pressing need for more objective and reliable diagnostic tools to ensure timely and accurate identification of ADHD.
While techniques like neuroimaging and electroencephalography can provide supplementary insights into brain activity patterns associated with attention deficits, their practical application in widespread ADHD diagnosis remains limited. These methods frequently demand specialized equipment, highly trained personnel, and substantial financial resources, creating barriers to access for many individuals and healthcare systems. Moreover, studies utilizing these technologies have often demonstrated only modest effect sizes, meaning the observed differences between individuals with and without ADHD are subtle and not always definitive. This necessitates cautious interpretation of results and highlights the ongoing need for more sensitive and readily available diagnostic tools that can complement, rather than rely solely upon, these complex and costly procedures.
The current diagnostic landscape for Attention-Deficit/Hyperactivity Disorder (ADHD) is significantly hampered by its reliance on behavioral assessments, which, while valuable, are inherently subjective and prone to variability. This necessitates the development of objective biomarkers – measurable indicators within the body – to improve diagnostic accuracy and facilitate earlier intervention. These biomarkers wouldn’t replace clinical evaluation, but rather serve as corroborating evidence, reducing misdiagnosis rates and enabling timely support for individuals struggling with ADHD. Identifying such reliable indicators represents a crucial step toward personalized medicine for ADHD, moving beyond symptom-based diagnosis to one informed by quantifiable biological data and potentially unlocking novel therapeutic targets. The pursuit of these biomarkers focuses on areas like genetic predispositions, neurochemical imbalances, and unique patterns of brain activity, promising a future where ADHD diagnosis is more precise, efficient, and ultimately, more effective.
Attention-Deficit/Hyperactivity Disorder (ADHD) is increasingly understood not simply as a behavioral condition, but as one rooted in metabolic dysfunction affecting brain function. Researchers posit that disruptions in how the body processes energy and nutrients – specifically involving neurotransmitter synthesis and neuronal energy production – contribute significantly to ADHD symptoms. Identifying reliable biomarkers, however, demands more than simply observing these metabolic shifts; it necessitates sophisticated analytical approaches. Advanced metabolomics, proteomics, and lipidomics – combined with machine learning algorithms – are crucial for discerning subtle yet significant patterns within biological samples. These robust analytical techniques can move beyond correlation to potentially reveal causal pathways, offering objective indicators for diagnosis and ultimately paving the way for personalized interventions targeting the underlying metabolic imbalances in ADHD.
Metabolic Fingerprints: A Deeper Signal
Metabolomic profiling involves the comprehensive identification and quantification of small molecule metabolites – including lipids, amino acids, and organic acids – within a biological sample. This approach differs from genomics or proteomics by directly measuring the end products of gene and protein activity, providing a snapshot of the organism’s physiological state. In the context of Attention-Deficit/Hyperactivity Disorder (ADHD), metabolomics allows researchers to identify disruptions in metabolic pathways, such as alterations in the concentrations of specific metabolites, which may serve as indicators of the disorder’s pathophysiology. By comparing the metabolic profiles of individuals with ADHD to control groups, researchers can pinpoint key pathways exhibiting significant differences, offering insights into the underlying biochemical processes associated with the condition and potential targets for therapeutic intervention.
Urine metabolomics offers a readily accessible and non-invasive biofluid for the assessment of metabolic alterations associated with Attention-Deficit/Hyperactivity Disorder (ADHD). Unlike methods requiring blood draws or cerebrospinal fluid analysis, urine collection is simple, repeatable, and suitable for diverse populations, including pediatric subjects. This ease of sample acquisition significantly reduces logistical challenges and costs associated with large-scale epidemiological studies and longitudinal research. Furthermore, urine contains a wealth of information reflecting systemic metabolism, providing a comprehensive snapshot of biochemical processes. The non-invasive nature of urine collection minimizes participant burden and facilitates the acquisition of repeated samples over time, crucial for tracking disease progression and treatment response.
Alterations in catecholamine metabolism, amino acid metabolism, and the urea cycle have been consistently observed in individuals with ADHD, indicating a broader systemic dysregulation beyond localized neurotransmitter imbalances. Specifically, studies reveal disrupted levels of dopamine and norepinephrine precursors and metabolites, alongside variations in the processing of essential amino acids like phenylalanine and tryptophan. Concurrent dysfunction of the urea cycle, responsible for ammonia detoxification, suggests potential impacts on brain energy metabolism and neurotransmitter synthesis. These interconnected metabolic disturbances highlight the complexity of ADHD pathophysiology and propose that metabolic profiling can identify systemic biomarkers reflecting the underlying biological processes.
The high dimensionality and inherent noise within metabolomic datasets necessitate the application of advanced machine learning techniques for effective biomarker discovery. Unsupervised methods, such as principal component analysis (PCA) and hierarchical clustering, are initially employed for data reduction and exploratory analysis, identifying patterns and potential groupings within the data. Subsequently, supervised learning algorithms – including support vector machines (SVM), random forests, and various regression models – are utilized to build predictive models and identify metabolites that significantly differentiate between control and ADHD groups. Feature selection methods are critical to minimize overfitting and prioritize biomarkers with robust predictive power. Cross-validation and independent cohort validation are essential steps to confirm the reliability and generalizability of identified biomarkers.
The Closest Resemblance: An Echo of Internal State
The Closest Resemblance (CR) classifier distinguishes itself in biomarker discovery through a combined methodological approach. It utilizes interval feature learning, a technique designed to address inherent biological variability by representing feature values as intervals rather than single points. This is coupled with an outranking-based similarity measure, which assesses the degree to which a sample’s feature intervals ‘outrank’ those of other samples, effectively quantifying their resemblance. This combination allows the classifier to identify biomarkers based on nuanced feature relationships and robustly handle the natural variation observed in biological data, rather than relying on precise, and potentially unstable, feature values.
Interval feature learning addresses inherent biological variability by representing biomarker concentrations not as single points, but as intervals reflecting the range of expected values for a given population or individual. This approach contrasts with traditional machine learning methods that often assume precise, fixed values, which can be unreliable due to individual differences and measurement error. By considering the uncertainty associated with biomarker levels, the classifier becomes less sensitive to outliers and more resilient to variations within and between subjects. Consequently, interval feature learning enhances the robustness of biomarker identification, leading to more reliable and generalizable results, even with limited sample sizes or noisy data.
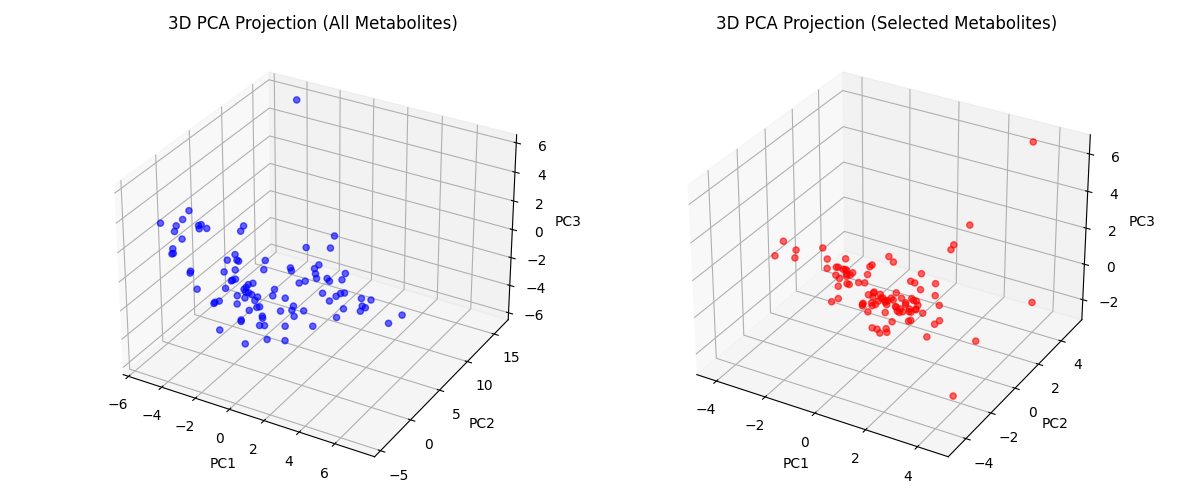
The biomarker set refinement process utilized greedy feature selection, a method that iteratively adds the metabolite providing the greatest discriminatory power, guided by Monte Carlo simulation. This simulation involved repeated random sampling with replacement from the original dataset to assess the stability and generalizability of each metabolite’s contribution to classification accuracy. By evaluating feature performance across multiple simulated datasets, the Monte Carlo approach mitigated the risk of selecting biomarkers that were merely artifacts of the specific training data. This process resulted in a highly refined biomarker set, ultimately reducing the initial pool of 60 urinary metabolites to a selection of only 14 for optimal classification performance, while maintaining high accuracy.
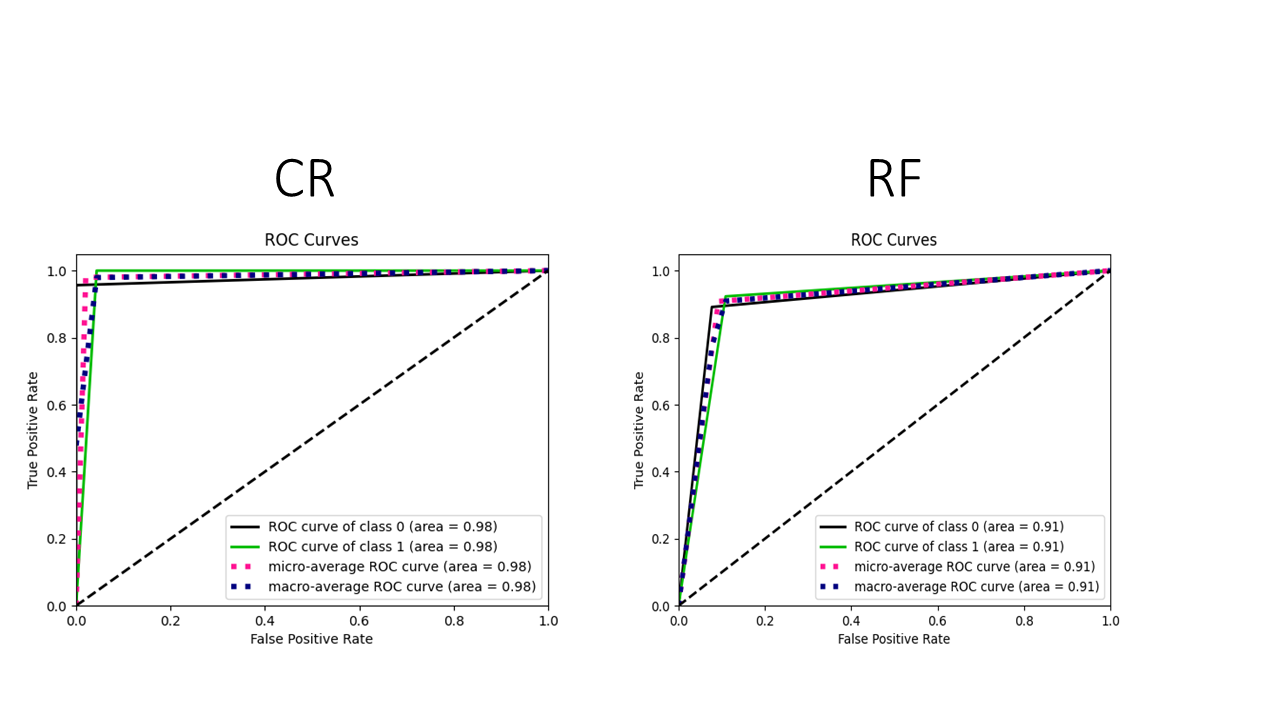
The Closest Resemblance (CR) classifier demonstrated 97.9% accuracy in differentiating between ADHD patients and control subjects, utilizing urinary metabolomics data and an integrated feature selection process. This performance was achieved with a highly refined biomarker set consisting of only 14 selected metabolites. This result indicates a substantial improvement over alternative machine learning algorithms; for example, a Random Forest classifier achieved only 91.8% accuracy when utilizing the complete set of 60 metabolites. The ability of the CR classifier to maintain high accuracy with a limited feature set suggests its potential for developing focused and efficient diagnostic tools.
Evaluation of the Closest Resemblance (CR) classifier demonstrated a 95.9% accuracy rate in distinguishing between ADHD patients and control subjects when utilizing the complete dataset of 60 urinary metabolites. This performance level, achieved without feature selection, indicates the inherent robustness of the CR classifier and its capacity to effectively discriminate between groups even with a larger, less refined feature set. While performance was further improved through feature selection to 97.9% with only 14 metabolites, the 95.9% accuracy using all 60 metabolites provides a strong baseline and suggests the classifier’s ability to leverage information across a broader metabolomic profile.
The Random Forest classifier, when applied to the complete urinary metabolomics dataset consisting of 60 metabolites, achieved an accuracy of 91.8%. This performance level serves as a benchmark for comparison with the Closest Resemblance (CR) classifier, which demonstrated superior results, attaining 95.9% accuracy with the same 60 metabolites and further improving to 97.9% accuracy when combined with an embedded feature selection process that reduced the metabolite set to 14.
Beyond Symptomology: Towards a Predictive Understanding
The pursuit of objective diagnostic tools for Attention-Deficit/Hyperactivity Disorder (ADHD) has traditionally relied on behavioral assessments, which can be subjective and prone to variability. However, recent research demonstrates the potential of integrating advanced machine learning with metabolomic profiling – the large-scale study of small molecules – to identify reliable biomarkers for the condition. This approach moves beyond symptom observation by analyzing the unique metabolic fingerprints present in individuals with ADHD, revealing underlying biochemical differences. Utilizing algorithms like the CR classifier, researchers can sift through complex metabolomic data, pinpointing specific metabolites that strongly correlate with ADHD diagnosis. This not only offers a pathway toward more accurate and consistent identification of the disorder, but also holds the promise of differentiating ADHD subtypes and understanding the biological mechanisms at play, ultimately improving diagnostic precision and paving the way for targeted interventions.
The potential for objective biomarkers in Attention-Deficit/Hyperactivity Disorder (ADHD) diagnosis represents a significant leap towards earlier and more accurate identification of the condition. Currently, diagnosis relies heavily on subjective behavioral assessments, which can be prone to inconsistencies and delays. The emergence of quantifiable biomarkers, derived from metabolomic profiling and refined by machine learning classifiers, offers the prospect of a more reliable and consistent diagnostic process. Earlier diagnosis, facilitated by these biomarkers, is critical because timely intervention – including behavioral therapies, educational support, and, when appropriate, medication – can substantially improve developmental trajectories and long-term outcomes for individuals with ADHD. By pinpointing the condition earlier, clinicians can implement tailored strategies to address core symptoms, mitigate associated challenges, and foster greater success in academic, social, and occupational settings, ultimately enhancing the quality of life for those affected.
The potential for personalized ADHD treatment stems from recognizing that metabolic dysregulation manifests uniquely in each individual. Rather than a one-size-fits-all approach, detailed metabolomic profiling can reveal specific biochemical imbalances contributing to a patient’s symptoms. This nuanced understanding allows clinicians to move beyond broad pharmacological interventions and tailor treatment plans – encompassing dietary adjustments, targeted supplementation, or optimized medication choices – to address the individual’s metabolic needs. By targeting the root biochemical disturbances, personalized strategies promise not only symptom management but also the potential to improve long-term outcomes and quality of life for those with ADHD, offering a future where treatment is as unique as the patient themselves.
The development of a computational classifier, known as CR, has yielded remarkably accurate results in distinguishing individuals with Attention-Deficit/Hyperactivity Disorder (ADHD) from neurotypical controls. Utilizing metabolomic data – a detailed biochemical profile of bodily fluids – the CR classifier achieved an area under the curve (AUC) of 0.98 when analyzing a streamlined selection of metabolites, and maintained a strong discriminatory power with an AUC of 0.96 when considering the full set of 60 metabolites. This near-perfect classification suggests a strong link between specific metabolic signatures and ADHD, hinting at the potential for objective, biologically-based diagnostic tools and moving beyond reliance on subjective behavioral assessments. The high AUC values demonstrate the classifier’s ability to correctly identify individuals with ADHD with exceptional accuracy, representing a significant step towards more reliable and earlier diagnosis.
Despite promising results demonstrating the potential of metabolomic profiling and machine learning in identifying objective ADHD biomarkers, substantial research remains essential before these findings can meaningfully impact clinical care. Rigorous validation studies, encompassing larger and more diverse patient cohorts, are needed to confirm the initial findings and establish the robustness of the identified metabolic signatures. Further investigation into the specific biological pathways disrupted in individuals with ADHD will refine understanding of the underlying disease mechanisms and inform the development of targeted therapies. Ultimately, translating these advancements into practical diagnostic tools and personalized treatment strategies requires collaborative efforts between researchers, clinicians, and regulatory bodies, with the shared goal of improving outcomes and quality of life for those affected by ADHD.
The pursuit of definitive biomarkers, as demonstrated by this study’s focus on urinary metabolites, echoes a timeless struggle against inherent system complexity. The Closest Resemblance classifier, with its embedded feature selection, attempts to impose order on a chaotic landscape of biochemical data. Yet, the very act of choosing ‘important’ features implies a frozen compromise, a prediction of future limitations. As Edsger W. Dijkstra observed, “It’s not about having the right tools, it’s about having the right perspective.” This work isn’t merely about finding biomarkers, but about acknowledging that any model-even one built with interpretable machine learning-is ultimately a simplification, a map that will inevitably diverge from the territory of the human condition.
What’s Next?
The pursuit of metabolomic signatures for ADHD diagnosis, as demonstrated by this work, is not a search for static truths, but the charting of a perpetually shifting landscape. Each identified metabolite, each optimized classifier, is merely a snapshot of a system in flux-a temporary reprieve from the inevitable drift towards false positives and diminishing returns. The Closest Resemblance method, while offering improved interpretability, does not abolish the fundamental problem: correlation is not causation, and the urinary metabolome reflects not just neurological state, but the totality of lived experience.
Future iterations will undoubtedly focus on expanding cohort sizes and incorporating longitudinal data. Yet, the real challenge lies not in accumulating more data points, but in accepting the inherent limitations of any predictive model. The system will find ways to confound the algorithm; dietary changes, environmental factors, even the very act of observation will introduce noise. Belief in a definitive biomarker panel is a denial of the body’s adaptive capacity.
The true metric of progress will not be accuracy scores, but the development of systems that gracefully degrade-that flag their own uncertainty, and adapt to the changing signatures of a complex disorder. It is a shift from seeking answers to embracing the questions, acknowledging that every diagnostic tool is, at its core, a temporary pact with chaos.
Original article: https://arxiv.org/pdf/2601.11283.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Clash Royale Best Boss Bandit Champion decks
- Vampire’s Fall 2 redeem codes and how to use them (June 2025)
- World Eternal Online promo codes and how to use them (September 2025)
- Best Arena 9 Decks in Clast Royale
- Country star who vanished from the spotlight 25 years ago resurfaces with viral Jessie James Decker duet
- ‘SNL’ host Finn Wolfhard has a ‘Stranger Things’ reunion and spoofs ‘Heated Rivalry’
- M7 Pass Event Guide: All you need to know
- Mobile Legends January 2026 Leaks: Upcoming new skins, heroes, events and more
- Solo Leveling Season 3 release date and details: “It may continue or it may not. Personally, I really hope that it does.”
- Kingdoms of Desire turns the Three Kingdoms era into an idle RPG power fantasy, now globally available
2026-01-19 19:35