Author: Denis Avetisyan
A new multi-agent framework dramatically boosts performance and efficiency in genomic question answering, pushing the boundaries of what’s possible with large language models.
GenomAgent achieves a 12% performance increase and a 79% reduction in computational cost compared to the original GeneGPT system on the GeneTuring benchmark.
Despite the increasing potential of large language models for knowledge-intensive tasks, effectively querying complex, distributed genomic databases remains a significant challenge. This work, ‘From Single to Multi-Agent Reasoning: Advancing GeneGPT for Genomics QA’, addresses this limitation by introducing GenomAgent, a novel multi-agent framework designed to surpass the performance of existing systems like GeneGPT. Through coordinated specialized agents, GenomAgent achieves a 12% average performance increase on the GeneTuring benchmark, alongside a substantial reduction in computational cost. Could this flexible, multi-agent architecture pave the way for more efficient knowledge extraction across diverse scientific disciplines?
The Inevitable Fragmentation of Genomic Inquiry
The pursuit of answers to intricate genomic questions is fundamentally hampered by the fragmented nature of biological data itself. Information relevant to a single query often resides across a multitude of databases – from gene expression profiles and protein interaction networks to scientific literature and clinical trial records – each employing different data formats, vocabularies, and levels of annotation. Traditional Natural Language Processing (NLP) methods, designed for more structured and homogenous text, struggle to effectively synthesize these disparate sources. The sheer heterogeneity necessitates complex data integration pipelines and sophisticated methods for resolving inconsistencies and ambiguities, creating a significant bottleneck in genomic research. Successfully navigating this challenge demands approaches capable of not only understanding individual data points, but also of reasoning across their varied contexts and representations to formulate coherent and accurate responses.
Current genomic question answering systems frequently falter when tasked with synthesizing information dispersed across multiple biological databases. These systems often exhibit limitations in their ability to perform the complex logical reasoning necessary to connect disparate data points, resulting in imprecise or incomplete answers. This poses a substantial hurdle for biological researchers, who rely on accurate and comprehensive information to drive discovery; a system that returns ambiguous results, or fails to integrate crucial evidence, diminishes its practical value and necessitates extensive manual verification. The inability to effectively navigate and reason over heterogeneous data sources, therefore, significantly restricts the broader applicability of these tools in accelerating biological understanding and translational research.
Recognizing the limitations of existing methods in tackling the complexities of genomic data, researchers turned to the development of advanced systems capable of robust and scalable question answering. This drive for improved performance initially led to the creation of GeneGPT, a large language model specifically adapted for genomic inquiries. However, acknowledging the need for even greater adaptability and comprehensive data integration, subsequent work focused on GenomAgent, a system designed to build upon GeneGPT’s foundation by incorporating agent-based reasoning and enhanced access to diverse biological databases. These tools represent a significant step towards automating complex genomic analyses and facilitating more efficient biological discovery, promising to unlock insights previously inaccessible due to the sheer volume and heterogeneity of available data.
GeneGPT: A Provisional Step Towards Intelligent Querying
GeneGPT is designed to respond directly to genomic queries without the need for extensive pre-training on specific datasets. This is achieved through in-context learning, where the large language model (LLM) is provided with examples of question-answer pairs and relevant genomic data within the prompt itself. Crucially, GeneGPT integrates with external Application Programming Interfaces (APIs) – such as those providing access to genomic databases – allowing the LLM to retrieve and utilize current information during the query process. This API integration, combined with the LLM’s inherent language processing capabilities, enables GeneGPT to synthesize information and formulate answers directly, functioning as a unified system for genomic question answering.
GeneGPT incorporates the ReAct (Reason + Act) framework to enhance its query processing capabilities. This framework enables the model to iteratively generate both reasoning traces and actions. During a query, ReAct allows GeneGPT to first reason about the information needed to answer the question, then decide on an appropriate action, such as an API call to a genomic database. The results of that action are then used to inform further reasoning, creating a cyclical process of thought and action. This contrasts with a direct question-answering approach by introducing a deliberate reasoning step and enabling dynamic action selection based on intermediate observations, ultimately improving the accuracy and reliability of responses to complex genomic inquiries.
GeneGPT’s initial implementation relied on a monolithic architecture, wherein all components – natural language processing, knowledge retrieval, and API interaction – were tightly integrated within a single system. This design, while functional for initial proof-of-concept, presented limitations regarding scalability to larger genomic datasets and the integration of novel data sources. Specifically, modifications to any component required complete retraining and redeployment of the entire model. Furthermore, complex queries demanding multiple iterative API calls and reasoning steps strained the monolithic structure, hindering performance and adaptability. These constraints motivated the development of a more modular and flexible architecture to facilitate independent component updates and improved handling of intricate genomic inquiries.
The functional architecture of GeneGPT incorporates a ‘Stop Token’ mechanism to reliably manage interactions with external Application Programming Interfaces (APIs). This token functions as a delimiter within the large language model’s generated text, signaling the conclusion of an API call and preventing the model from continuing to generate text that would be misinterpreted as further API instructions. Specifically, when GeneGPT generates the Stop Token during the ReAct process, it halts the API call sequence and returns control to the language model for subsequent reasoning or action selection. This ensures that API calls are properly terminated, avoiding errors or unintended consequences stemming from incomplete or erroneous API requests and contributing to the overall stability of the query process.
GenomAgent: A Multi-Agent Framework for Robust Querying
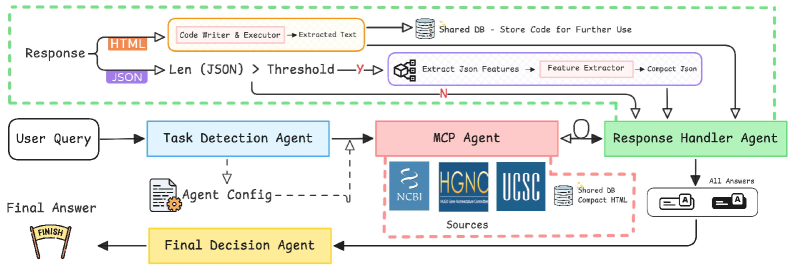
GenomAgent employs a multi-agent architecture to address complex genomic queries by dividing the question-answering process into discrete, specialized agents. Each agent is designed to perform a specific task, such as identifying the query’s intent, retrieving relevant data from databases, extracting key features, writing code for data manipulation, or formatting the final response. This decomposition allows for parallel processing and modularity, enhancing the system’s ability to handle varied query types and data sources. By assigning responsibility for individual tasks to dedicated agents, GenomAgent facilitates a more robust and scalable approach to genomic information retrieval compared to monolithic systems.
The GenomAgent framework utilizes a collaborative process between specialized agents, notably the Task Detection Agent and the Code Writer Agent, to query and integrate data from heterogeneous biological databases. The Task Detection Agent analyzes incoming queries to determine the necessary data retrieval steps, while the Code Writer Agent dynamically generates and executes database-specific queries – currently supporting NCBI, HGNC, and UCSC – to access relevant information. This interaction is governed by the Multi-source Coordination Protocol, which manages data requests, handles potential conflicts arising from differing database schemas, and ensures consistent data formatting before passing the results to subsequent agents for processing and response generation.
The GenomAgent framework incorporates dedicated agents to manage data integrity and output coherence. The ‘Feature Extractor Agent’ rigorously processes retrieved data, identifying and correcting inconsistencies or errors in feature representation. Subsequently, the ‘Response Handler Agent’ standardizes data formats to ensure compatibility and facilitates consistent presentation of information. Finally, the ‘Final Decision Agent’ integrates the processed and formatted outputs from all preceding agents, applying a synthesis process to generate a single, comprehensive, and logically structured response to the initial query.
GenomAgent leverages the Google Agent Development Kit (GAK) to facilitate a highly modular architecture. This foundation enables the straightforward integration of diverse data streams – from updated genomic databases to emerging proteomic analyses – without requiring substantial code refactoring. Agents can be readily adapted or replaced to accommodate differing data schemas and access protocols. Similarly, the framework supports the addition of new query types through the creation of specialized agents or the modification of existing agent behaviors. The GAK provides tools for inter-agent communication and orchestration, simplifying the process of incorporating new functionalities and ensuring seamless integration within the existing multi-agent system. This modular design promotes scalability and adaptability, allowing GenomAgent to evolve with changing data landscapes and user requirements.
Validating Performance and Ensuring Reproducibility
The GeneTuring Benchmark is designed as a controlled and consistent evaluation environment for large language models applied to genomic tasks. It provides a defined set of challenges and metrics, enabling quantitative comparison of model performance. This benchmark facilitates the assessment of both GeneGPT and GenomAgent across a standardized dataset, isolating the impact of architectural or algorithmic modifications. The framework’s consistent methodology allows for reproducible results and objective measurement of improvements in areas such as gene function prediction, variant effect analysis, and other critical genomic tasks, moving beyond subjective assessments of performance.
Reproducibility of results and assessment of architectural impact were addressed through recent studies leveraging large language models and software frameworks. Specifically, GPT-4o-mini was employed as a testing platform, and the LangGraph framework facilitated the construction and execution of complex, multi-step reasoning processes. This allowed for systematic variation of system architecture and detailed tracking of performance metrics, ensuring that observed improvements were attributable to specific design choices and not random fluctuations. The controlled environment enabled consistent evaluation across different configurations, bolstering confidence in the system’s stability and facilitating iterative refinement.
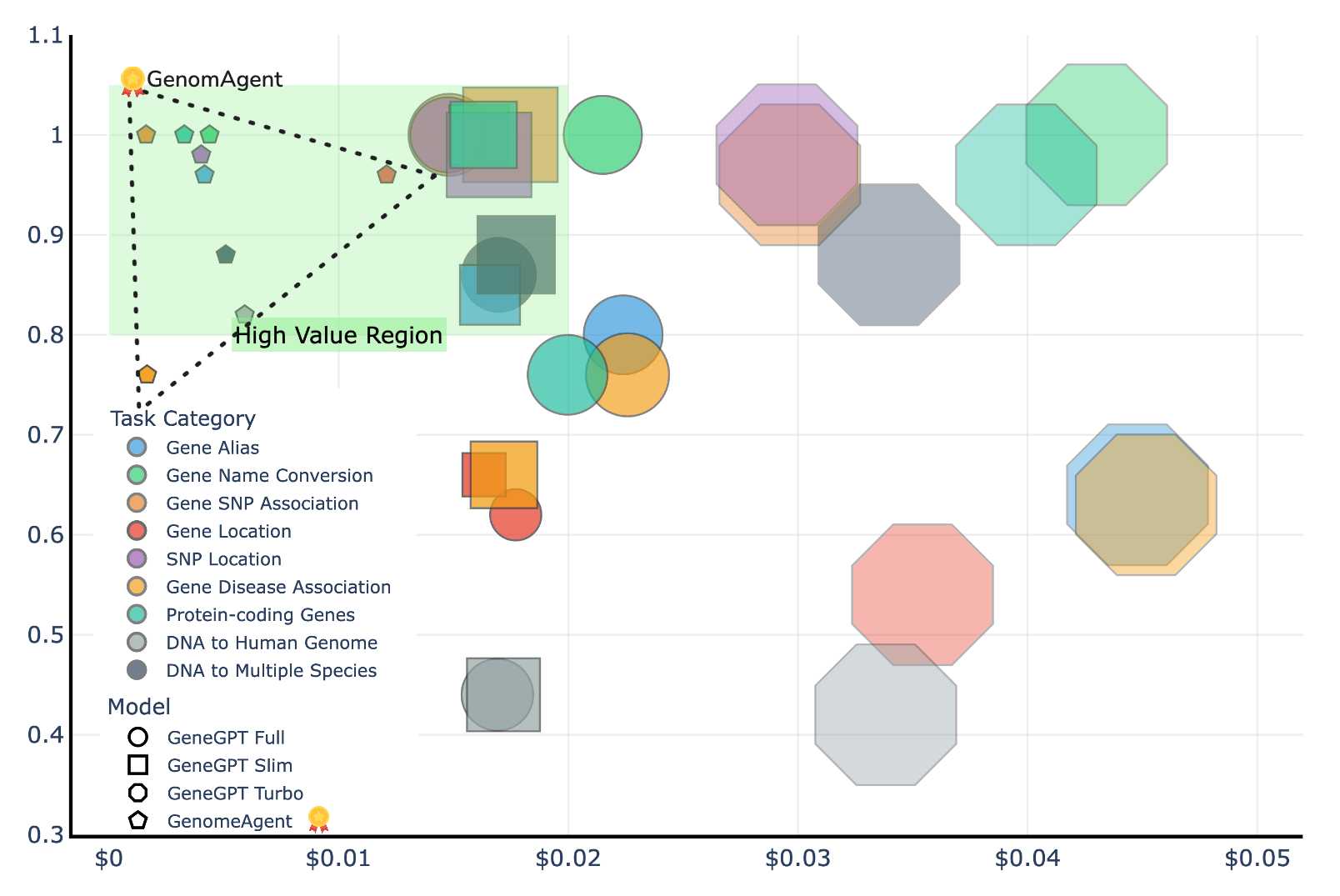
Evaluations conducted using the GeneTuring benchmark demonstrate a quantifiable performance advantage for GenomAgent over GeneGPT. Specifically, GenomAgent achieved an average performance score of 0.93, representing a 12% increase compared to GeneGPT’s score of 0.83. Furthermore, GenomAgent exhibited a 79% reduction in computational cost, totaling \$10.06, indicating a significantly more efficient operational expense compared to GeneGPT. These metrics were consistently observed during testing on the benchmark and provide objective data regarding the relative capabilities of each system.
Evaluations conducted using the GeneTuring Benchmark demonstrate that GenomAgent achieves a 28.8% performance increase on alignment tasks compared to GeneGPT. Alignment tasks, within the benchmark, assess the model’s ability to correctly map genetic sequences to their corresponding functions or phenotypes. This improvement indicates that GenomAgent more accurately predicts the functional consequences of genetic variations than GeneGPT, suggesting enhanced capabilities in tasks such as gene annotation and variant effect prediction. The metric used for this evaluation is not specified, but the reported percentage difference represents a statistically significant improvement in performance on these specific alignment challenges.
Systematic evaluation and refinement are foundational to the development of reliable AI systems for genomic research. Rigorous benchmarking, such as that provided by the GeneTuring benchmark, allows for quantifiable performance assessment and identification of areas for improvement. Data from recent studies demonstrate that GenomAgent, through iterative refinement based on benchmark results, achieves a 12% performance increase and a 79% reduction in computational cost compared to GeneGPT. This data-driven approach not only validates the efficacy of architectural changes but also fosters confidence in the system’s outputs, which is essential for accelerating scientific discovery and enabling broader adoption of AI-driven genomic tools.
Future Directions: Towards Intelligent Genomic Exploration
The core of GenomAgent’s potential lies in its deliberately constructed multi-agent architecture, designed not as a rigid system, but as a highly adaptable framework. This approach facilitates the seamless incorporation of diverse data streams – from updated genomic databases to emerging proteomic analyses – without requiring fundamental code restructuring. Furthermore, the modular design allows for the integration of novel computational models, whether machine learning algorithms predicting gene function or statistical methods assessing data significance. Crucially, the system’s reasoning capabilities can be readily extended through the addition of specialized agents focused on specific analytical tasks, creating a dynamically evolving platform capable of addressing increasingly complex biological questions and adapting to the ever-expanding landscape of genomic knowledge.
Ongoing development centers on refining the agents’ capacity to navigate the inherent intricacies of genomic data, specifically addressing complex interdependencies between genes, proteins, and pathways. This involves equipping them with advanced reasoning mechanisms to interpret incomplete or noisy datasets-a common challenge in biological research-and to dynamically adjust analytical strategies in response to evolving queries. Researchers aim to move beyond static analyses, enabling the system to learn from incoming data and refine its hypotheses in real-time, ultimately fostering a more adaptive and insightful exploration of the genome and accelerating the identification of meaningful biological patterns.
The enhanced capabilities promise a paradigm shift in genomic research, allowing scientists to navigate the immense complexity of biological data with remarkable speed and precision. By automating and optimizing the process of data analysis, researchers can significantly reduce the time required to identify meaningful patterns and correlations – insights previously obscured by the sheer volume of information. This accelerated discovery cycle isn’t merely about faster processing; it’s about unlocking a deeper understanding of fundamental biological processes, potentially leading to breakthroughs in areas like personalized medicine, disease prevention, and the development of novel therapies. The capacity to uncover previously hidden connections within genomic data represents a powerful tool for advancing the frontiers of biological knowledge and translating those discoveries into real-world applications.
The ultimate aim of this research extends beyond current genomic analysis tools, envisioning a system capable of facilitating truly intelligent exploration of biological data. This future system isn’t simply about faster processing or larger datasets; it’s about empowering scientists to formulate increasingly sophisticated questions-questions currently beyond the reach of conventional methods-and receive meaningful, data-driven answers. By integrating advanced multi-agent reasoning and adaptive learning, the system anticipates a shift from hypothesis-driven research to a more exploratory approach, where unexpected patterns and novel connections within genomic data are readily revealed, ultimately accelerating the pace of biological discovery and fostering a deeper understanding of life’s complexities.
The pursuit of increasingly capable systems, such as GeneGPT and now GenomAgent, reveals a fundamental truth about software architecture: initial designs are rarely sufficient for long-term viability. GenomAgent’s 12% performance increase and 79% reduction in computational cost aren’t merely incremental gains, but evidence of adaptation-a crucial element for graceful aging. As Barbara Liskov observed, “It’s one of the hardest things to do: to design something that can change.” This sentiment perfectly encapsulates the shift from a single-agent to a multi-agent framework; it isn’t about replacing the old, but building upon it with a system designed to accommodate future evolution and the inevitable expansion of genomic knowledge.
The Long View
The pursuit of increasingly capable question-answering systems in genomics, as demonstrated by this work, inevitably confronts a fundamental truth: systems learn to age gracefully. While a 12% performance increase and 79% reduction in computational cost represent tangible progress, they are merely points along a curve of diminishing returns. The underlying complexities of biological systems, and the inherent limitations of any model attempting to represent them, will always present a moving target. The focus shifts, then, from simply achieving higher scores on benchmarks to understanding how these systems fail, and what those failures reveal about the limits of current approaches.
The integration of multiple agents, as showcased by GenomAgent, is a logical progression, mirroring the distributed nature of biological processes themselves. However, true intelligence isn’t solely about speed or accuracy; it’s about efficient resource allocation and adaptability. The question isn’t whether a system can answer more questions, but whether it can learn to ask better ones, and to recognize when an answer is beyond its grasp.
Sometimes observing the process is better than trying to speed it up. The GeneTuring benchmark, and others like it, serve as useful diagnostic tools, but they shouldn’t be mistaken for destinations. The field will likely move toward more nuanced evaluation metrics, and a greater emphasis on systems that can not only provide answers, but also articulate their own uncertainties and limitations.
Original article: https://arxiv.org/pdf/2601.10581.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Clash Royale Best Boss Bandit Champion decks
- Vampire’s Fall 2 redeem codes and how to use them (June 2025)
- World Eternal Online promo codes and how to use them (September 2025)
- Best Arena 9 Decks in Clast Royale
- How to find the Roaming Oak Tree in Heartopia
- Country star who vanished from the spotlight 25 years ago resurfaces with viral Jessie James Decker duet
- Mobile Legends January 2026 Leaks: Upcoming new skins, heroes, events and more
- M7 Pass Event Guide: All you need to know
- Solo Leveling Season 3 release date and details: “It may continue or it may not. Personally, I really hope that it does.”
- Kingdoms of Desire turns the Three Kingdoms era into an idle RPG power fantasy, now globally available
2026-01-17 17:05